Generative artificial intelligence enables the generation of bone scintigraphy images and improves generalization of deep learning models in data-constrained environments
- PMID: 39878897
- PMCID: PMC12119683
- DOI: 10.1007/s00259-025-07091-8
Generative artificial intelligence enables the generation of bone scintigraphy images and improves generalization of deep learning models in data-constrained environments
Abstract
Purpose: Advancements of deep learning in medical imaging are often constrained by the limited availability of large, annotated datasets, resulting in underperforming models when deployed under real-world conditions. This study investigated a generative artificial intelligence (AI) approach to create synthetic medical images taking the example of bone scintigraphy scans, to increase the data diversity of small-scale datasets for more effective model training and improved generalization.
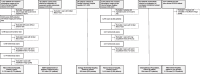
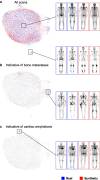
Methods: We trained a generative model on 99mTc-bone scintigraphy scans from 9,170 patients in one center to generate high-quality and fully anonymized annotated scans of patients representing two distinct disease patterns: abnormal uptake indicative of (i) bone metastases and (ii) cardiac uptake indicative of cardiac amyloidosis. A blinded reader study was performed to assess the clinical validity and quality of the generated data. We investigated the added value of the generated data by augmenting an independent small single-center dataset with synthetic data and by training a deep learning model to detect abnormal uptake in a downstream classification task. We tested this model on 7,472 scans from 6,448 patients across four external sites in a cross-tracer and cross-scanner setting and associated the resulting model predictions with clinical outcomes.
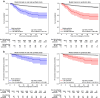
Results: The clinical value and high quality of the synthetic imaging data were confirmed by four readers, who were unable to distinguish synthetic scans from real scans (average accuracy: 0.48% [95% CI 0.46-0.51]), disagreeing in 239 (60%) of 400 cases (Fleiss' kappa: 0.18). Adding synthetic data to the training set improved model performance by a mean (± SD) of 33(± 10)% AUC (p < 0.0001) for detecting abnormal uptake indicative of bone metastases and by 5(± 4)% AUC (p < 0.0001) for detecting uptake indicative of cardiac amyloidosis across both internal and external testing cohorts, compared to models without synthetic training data. Patients with predicted abnormal uptake had adverse clinical outcomes (log-rank: p < 0.0001).
Conclusions: Generative AI enables the targeted generation of bone scintigraphy images representing different clinical conditions. Our findings point to the potential of synthetic data to overcome challenges in data sharing and in developing reliable and prognostic deep learning models in data-limited environments.
Keywords: Artificial intelligence; Bone scintigraphy; Generative; Multicenter; Synthetic data.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors. Consent to participate: For the patient cohort from Careggi University Hospital, the patient’s signature on the examination authorization form included consent for the use of anonymized data for research purposes. The patient cohort from West China Hospital is an open-access dataset. For all other cohorts, the requirement to obtain informed consent was waived. Competing interests: Christian Nitsche reports speaker fees or institutional research grants from Pfizer and advisory board honoraria from Prothena.
Figures


References
-
- Spielvogel CP, Haberl D, Mascherbauer K, et al. Diagnosis and prognosis of abnormal cardiac scintigraphy uptake suggestive of cardiac amyloidosis using artificial intelligence: a retrospective, international, multicentre, cross-tracer development and validation study. Lancet Digit Health. 2024;6:e251–60. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

