Recalibrating Protection Factors Using Millisecond Hydrogen/Deuterium Exchange Mass Spectrometry
- PMID: 39879324
- PMCID: PMC11822740
- DOI: 10.1021/acs.analchem.4c03631
Recalibrating Protection Factors Using Millisecond Hydrogen/Deuterium Exchange Mass Spectrometry
Abstract
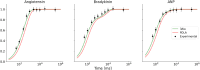
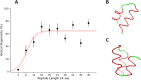
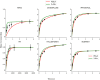
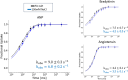
Hydrogen/deuterium exchange mass spectrometry (HDX-MS) is a powerful technique to interrogate protein structure and dynamics. With the ability to study almost any protein without a size limit, including intrinsically disordered ones, HDX-MS has shown fast growing importance as a complement to structural elucidation techniques. Current experiments compare two or more related conditions (sequences, interaction partners, excipients, conformational states, etc.) to determine statistically significant differences at a number of fixed time points and highlight areas of changed structural dynamics in the protein. The work presented here builds on the fundamental research performed in the early days of the technique and re-examines exchange rate calculations with the aim of establishing HDX-MS as an absolute and quantitative, rather than relative and qualitative, measurement. We performed millisecond HDX-MS experiments on a mixture of three unstructured peptides (angiotensin, bradykinin, and atrial natriuretic peptide amide rat) and compared experimental deuterium uptake curves with theoretical ones predicted using established exchange rate calculations. With poly-dl-alanine (PDLA) commonly used as a reference,, we find that experimental rates are sometimes faster than theoretically possible, while they agree much better, and are never faster, with the fully unstructured trialanine peptide (3-Ala). Molecular dynamics (MD) simulations confirm the high helical propensity of the longer and partially structured PDLA peptides, which need as few as 15 residues to form a stable helix and are therefore not suitable as an unstructured reference. Reanalysis of previously published data by Weis et al. at 100 mM NaCl however still shows a discrepancy with predictions based on 3-Ala in the absence of salt, highlighting the need for a better understanding of salt effects on exchange rates. Such currently unquantifiable salt effects prevent us from proposing a comprehensive, universal calibration framework at the moment. Nevertheless, an accurate recalibration of intrinsic exchange rate calculations is crucial to enable kinetic modeling of the exchange process and to ultimately allow HDX-MS to move toward a direct link with atomistic structural models.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Solid-State Hydrogen-Deuterium Exchange Mass Spectrometry (ssHDX-MS) of Lyophilized Poly-d,l-Alanine.Mol Pharm. 2019 Jul 1;16(7):2935-2946. doi: 10.1021/acs.molpharmaceut.9b00162. Epub 2019 Jun 18. Mol Pharm. 2019. PMID: 31244225
-
Online Fully Automated System for Hydrogen/Deuterium-Exchange Mass Spectrometry with Millisecond Time Resolution.Anal Chem. 2023 Mar 21;95(11):5000-5008. doi: 10.1021/acs.analchem.2c05310. Epub 2023 Mar 9. Anal Chem. 2023. PMID: 36896500 Free PMC article.
-
A Hydrogen-Deuterium Exchange Mass Spectrometry (HDX-MS) Platform for Investigating Peptide Biosynthetic Enzymes.J Vis Exp. 2020 May 4;(159). doi: 10.3791/61053. J Vis Exp. 2020. PMID: 32420996
-
Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling.Acc Chem Res. 2014 Oct 21;47(10):3018-27. doi: 10.1021/ar500194w. Epub 2014 Aug 29. Acc Chem Res. 2014. PMID: 25171396 Review.
-
Bridging protein structure, dynamics, and function using hydrogen/deuterium-exchange mass spectrometry.Protein Sci. 2020 Apr;29(4):843-855. doi: 10.1002/pro.3790. Epub 2019 Nov 25. Protein Sci. 2020. PMID: 31721348 Free PMC article. Review.
References
-
- Theillet F.-X.; Binolfi A.; Frembgen-Kesner T.; Hingorani K.; Sarkar M.; Kyne C.; Li C.; Crowley P. B.; Gierasch L.; Pielak G. J.; Elcock A. H.; Gershenson A.; Selenko P. Physicochemical Properties of Cells and Their Effects on Intrinsically Disordered Proteins (IDPs). Chem. Rev. 2014, 114 (13), 6661–6714. 10.1021/cr400695p. - DOI - PMC - PubMed
-
- Masson G. R.; Burke J. E.; Ahn N. G.; Anand G. S.; Borchers C.; Brier S.; Bou-Assaf G. M.; Engen J. R.; Englander S. W.; Faber J.; Garlish R.; Griffin P. R.; Gross M. L.; Guttman M.; Hamuro Y.; Heck A. J. R.; Houde D.; Iacob R. E.; Jørgensen T. J. D.; Kaltashov I. A.; Klinman J. P.; Konermann L.; Man P.; Mayne L.; Pascal B. D.; Reichmann D.; Skehel M.; Snijder J.; Strutzenberg T. S.; Underbakke E. S.; Wagner C.; Wales T. E.; Walters B. T.; Weis D. D.; Wilson D. J.; Wintrode P. L.; Zhang Z.; Zheng J.; Schriemer D. C.; Rand K. D. Recommendations for Performing, Interpreting and Reporting Hydrogen Deuterium Exchange Mass Spectrometry (HDX-MS) Experiments. Nat. Methods 2019, 16 (7), 595–602. 10.1038/s41592-019-0459-y. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

