Sequence variants in HECTD1 result in a variable neurodevelopmental disorder
- PMID: 39879987
- PMCID: PMC11947180
- DOI: 10.1016/j.ajhg.2025.01.001
Sequence variants in HECTD1 result in a variable neurodevelopmental disorder
Abstract
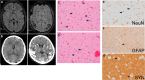
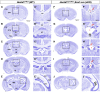
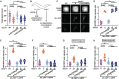
Dysregulation of genes encoding the homologous to E6AP C-terminus (HECT) E3 ubiquitin ligases has been linked to cancer and structural birth defects. One member of this family, the HECT-domain-containing protein 1 (HECTD1), mediates developmental pathways, including cell signaling, gene expression, and embryogenesis. Through GeneMatcher, we identified 14 unrelated individuals with 15 different variants in HECTD1 (10 missense, 3 frameshift, 1 nonsense, and 1 splicing variant) with neurodevelopmental disorders (NDDs), including autism, attention-deficit/hyperactivity disorder, and epilepsy. Of these 15 HECTD1 variants, 10 occurred de novo, 3 had unknown inheritance, and 2 were compound heterozygous. While all individuals in this cohort displayed NDDs, no genotype-phenotype correlation was apparent. Conditional knockout of Hectd1 in the neural lineage in mice resulted in microcephaly, severe hippocampal malformations, and complete agenesis of the corpus callosum, supporting a role for Hectd1 in embryonic brain development. Functional studies of select variants in C. elegans revealed dominant effects, including either change-of-function or loss-of-function/haploinsufficient mechanisms, which may explain phenotypic heterogeneity. Significant enrichment of de novo variants in HECTD1 was also shown in an independent cohort of 53,305 published trios with NDDs or congenital heart disease. Thus, our clinical and functional data support a critical requirement of HECTD1 for human brain development.
Keywords: HECTD1; autism; epilepsy; neurodevelopmental disorders; ubiquitin-proteasome system.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.G.M. and M.J.G.S. are employees of GeneDx, LLC.
Figures

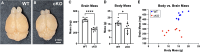
References
-
- Ebstein F., Küry S., Papendorf J.J., Krüger E. Neurodevelopmental Disorders (NDD) Caused by Genomic Alterations of the Ubiquitin-Proteasome System (UPS): the Possible Contribution of Immune Dysregulation to Disease Pathogenesis. Front. Mol. Neurosci. 2021;14 doi: 10.3389/fnmol.2021.733012. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

