Low-coverage whole genome sequencing of low-grade dysplasia strongly predicts advanced neoplasia risk in ulcerative colitis
- PMID: 39880602
- PMCID: PMC12013573
- DOI: 10.1136/gutjnl-2024-333353
Low-coverage whole genome sequencing of low-grade dysplasia strongly predicts advanced neoplasia risk in ulcerative colitis
Abstract
Background: The risk of developing advanced neoplasia (AN; colorectal cancer and/or high-grade dysplasia) in ulcerative colitis (UC) patients with a low-grade dysplasia (LGD) lesion is variable and difficult to predict. This is a major challenge for effective clinical management.
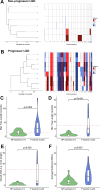
Objective: We aimed to provide accurate AN risk stratification in UC patients with LGD. We hypothesised that the pattern and burden of somatic genomic copy number alterations (CNAs) in LGD lesions could predict future AN risk.
Design: We performed a retrospective multicentre validated case-control study using n=270 LGD samples from n=122 patients with UC. Patients were designated progressors (n=40) if they had a diagnosis of AN in the ~5 years following LGD diagnosis or non-progressors (n=82) if they remained AN-free during follow-up. DNA was extracted from the baseline LGD lesion, low-coverage whole genome sequencing performed and data processed to detect CNAs. Survival analysis was used to evaluate CNAs as predictors of future AN risk.
Results: CNA burden was significantly higher in progressors than non-progressors (p=2×10-6 in discovery cohort) and was a very significant predictor of AN risk in univariate analysis (OR=36; p=9×10-7), outperforming existing clinical risk factors such as lesion size, shape and focality. Optimal risk prediction was achieved with a multivariate model combining CNA burden with the known clinical risk factor of incomplete LGD resection. Within-LGD lesion genetic heterogeneity did not confound risk prediction.
Conclusion: Measurement of CNAs in LGD is an accurate predictor of AN risk in inflammatory bowel disease and is likely to support clinical management.
Keywords: CANCER PREVENTION; COLORECTAL CANCER; GENETIC INSTABILITY; INFLAMMATORY BOWEL DISEASE; ULCERATIVE COLITIS.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: The authors are in discussions about potential commercialisation and clinical translation of the findings described here. AH has served as consultant, advisory board member or speaker for AbbVie, Arena, Atlantic, Bristol-Myers Squibb, Celgene, Celltrion, Falk, Galapogos, Lilly, Janssen, MSD, Napp Pharmaceuticals, Pfizer, Pharmacosmos, Roche, Shire and Takeda. KC has an investigator-led research grant from Phathom Pharmaceuticals. TAG and A-MB are named as coinventors on a patent application that describe a method for TCR sequencing (GB2305655.9), and TAG is named on a method to measure evolutionary dynamics in cancers using DNA methylation (GB2317139.0). TAG has received honorarium from Genentech and DAiNA therapeutics.
Figures




Update of
-
Low coverage whole genome sequencing of low-grade dysplasia strongly predicts colorectal cancer risk in ulcerative colitis.medRxiv [Preprint]. 2024 Jul 8:2024.07.08.24309811. doi: 10.1101/2024.07.08.24309811. medRxiv. 2024. Update in: Gut. 2025 Apr 7;74(5):740-751. doi: 10.1136/gutjnl-2024-333353. PMID: 39040198 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
