Molecular and pharmacological heterogeneity of ETV6::RUNX1 acute lymphoblastic leukemia
- PMID: 39880832
- PMCID: PMC11779914
- DOI: 10.1038/s41467-025-56229-7
Molecular and pharmacological heterogeneity of ETV6::RUNX1 acute lymphoblastic leukemia
Abstract
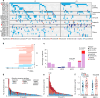
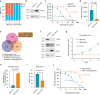
ETV6::RUNX1 is the most common fusion gene in childhood acute lymphoblastic leukemia (ALL) associated with favorable prognosis, but the optimal therapy for this subtype remains unclear. Profiling the genomic and pharmacological landscape of 194 pediatric ETV6::RUNX1 ALL cases, we uncover two transcriptomic clusters, C1 (61%) and C2 (39%). Compared to C1, the C2 subtype features higher white blood cell counts and younger age at diagnosis, as well as better early treatment responses. Pharmacologically, C2 is more sensitive to thiopurines and prednisolone, partially explained by the enrichment of PAX5 deletions. Re-introducing PAX5 in ETV6::RUNX1 ALL of the C2 subtype converts its gene expression and drug resistance profile to C1, with partial blockade of G1 to S transition mediated by CDK6 expression. Our results point to molecular heterogeneity within ETV6::RUNX1 ALL linked to divergent drug responses, providing insights into the pathogenesis and therapeutic vulnerability of this common pediatric ALL subtype.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Mullighan, C. G. The molecular genetic makeup of acute lymphoblastic leukemia. Hematol. Am. Soc. Hematol. Educ. Program10.1182/asheducation-2012.1.389 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

