VSTM2L protects prostate cancer cells against ferroptosis via inhibiting VDAC1 oligomerization and maintaining mitochondria homeostasis
- PMID: 39880844
- PMCID: PMC11779845
- DOI: 10.1038/s41467-025-56494-6
VSTM2L protects prostate cancer cells against ferroptosis via inhibiting VDAC1 oligomerization and maintaining mitochondria homeostasis
Abstract
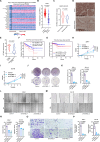
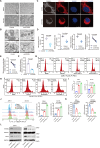
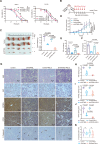
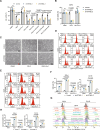
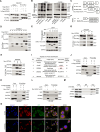
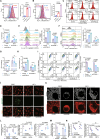
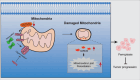
Ferroptosis is a form of iron-dependent programmed cell death, which is distinct from apoptosis, necrosis, and autophagy. Mitochondria play a critical role in initiating and amplifying ferroptosis in cancer cells. Voltage-Dependent Anion Channel 1 (VDAC1) embedded in the mitochondrial outer membrane, exerts roles in regulation of ferroptosis. However, the mechanisms of VDAC1 oligomerization in regulating ferroptosis are not well elucidated. Here, we identify that a VDAC1 binding protein V-Set and Transmembrane Domain Containing 2 Like (VSTM2L), mainly localized to mitochondria, is positively associated with prostate cancer (PCa) progression, and a key regulator of ferroptosis. Moreover, VSTM2L knockdown in PCa cells enhances the sensitivity of RSL3-induced ferroptosis. Mechanistically, VSTM2L forms complex with VDAC1 and hexokinase 2 (HK2), enhancing their binding affinity and preventing VDAC1 oligomerization, thereby inhibiting ferroptosis and maintaining mitochondria homeostasis in vitro and in vivo. Collectively, our findings reveal a pivotal role for mitochondria-localized VSTM2L in driving ferroptosis resistance and highlight its potential as a ferroptosis-inducing therapeutic target for the treatment of PCa.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clinicians71, 209–249 (2021). - PubMed
-
- Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA: A Cancer J. Clinicians74, 12–49 (2024). - PubMed
-
- Pernigoni, N. et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science374, 216–224 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

