CARD domains mediate anti-phage defence in bacterial gasdermin systems
- PMID: 39880956
- PMCID: PMC12373157
- DOI: 10.1038/s41586-024-08498-3
CARD domains mediate anti-phage defence in bacterial gasdermin systems
Abstract
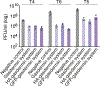
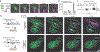
Caspase recruitment domains (CARDs) and pyrin domains are important facilitators of inflammasome activity and pyroptosis1. Following pathogen recognition by nucleotide binding-domain, leucine-rich, repeat-containing (NLR) proteins, CARDs recruit and activate caspases, which, in turn, activate gasdermin pore-forming proteins to induce pyroptotic cell death2. Here we show that CARD domains are present in defence systems that protect bacteria against phage. The bacterial CARD domain is essential for protease-mediated activation of certain bacterial gasdermins, which promote cell death once phage infection is recognized. We further show that multiple anti-phage defence systems use CARD domains to activate a variety of cell death effectors, and that CARD domains mediate protein-protein interactions in these systems. We find that these systems are triggered by a conserved immune-evasion protein used by phages to overcome the bacterial defence system RexAB3, demonstrating that phage proteins inhibiting one defence system can activate another. Our results suggest that CARD domains represent an ancient component of innate immune systems conserved from bacteria to humans, and that CARD-dependent activation of gasdermins is shared in organisms across the tree of life.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: R.S. is a scientific cofounder and advisor of BiomX and Ecophage. The other authors declare no competing interests.
Figures


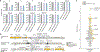
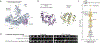






Update of
-
CARD-like domains mediate anti-phage defense in bacterial gasdermin systems.bioRxiv [Preprint]. 2023 May 29:2023.05.28.542683. doi: 10.1101/2023.05.28.542683. bioRxiv. 2023. Update in: Nature. 2025 Mar;639(8055):727-734. doi: 10.1038/s41586-024-08498-3. PMID: 37398489 Free PMC article. Updated. Preprint.
References
-
- Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). - PubMed
-
- Wong S, Alattas H & Slavcev RA A snapshot of the λ T4rII exclusion (Rex) phenotype in Escherichia coli. Curr. Genet. 67, 739–745 (2021). - PubMed
-
- Broz P, Pelegrín P & Shao F The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20, 143–157 (2020). - PubMed
Additional references
-
- Millman A et al. Bacterial Retrons Function In Anti-Phage Defense. Cell 183, 1551–1561.e12 (2020). - PubMed
-
- Mazzocco A, Waddell TE, Lingohr E, and Johnson RP Enumeration of bacteriophages using the small drop plaque assay system. Bacteriophages: Methods and Protocols (Humana Press; ), 81–85 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

