Subtypes A1 and D, and recombinant HIV-1 natural polymorphisms associated with lenacapavir drug resistance in Uganda
- PMID: 39881519
- PMCID: PMC11962371
- DOI: 10.1093/jac/dkaf018
Subtypes A1 and D, and recombinant HIV-1 natural polymorphisms associated with lenacapavir drug resistance in Uganda
Abstract
Background: Lenacapavir, a novel HIV-1 capsid inhibitor, shows promise for treating MDR HIV-1, as well as for pre-exposure prophylaxis (PrEP) in prevention of HIV infection. Its unique mechanism and lack of cross-resistance with other antiretroviral classes make lenacapavir a significant addition to HIV therapy. The clinical trials CALIBRATE and CAPELLA have demonstrated high viral suppression rates in both ART-naive individuals and individuals with MDR HIV-1. Lenacapavir-associated resistance mutations, such as M66I and Q67H, rarely seen as natural polymorphisms in lenacapavir-naive populations, are predominantly studied in subtype B HIV-1.
Objectives: Our study aimed to investigate the prevalence of lenacapavir resistance-associated mutations in HIV-1 subtypes A1 and D in a cohort of individuals living with HIV-1 from southwestern Uganda.
Methods: Utilizing plasma samples from ART-naive adults living in Uganda, HIV-1 Gag p24 (capsid) sequences were analysed for lenacapavir resistance mutations.
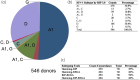
Results: Among 546 lenacapavir-naive participants, no major lenacapavir resistance-associated mutations were found. Minor mutations were present in 1.6% of participants, with T107A being the most common. Longitudinal data indicated the persistence of T107A for at least 3 years post-ART initiation in one participant. Phylogenetic analysis indicated individuals carrying T107A were found independently in distinct locations within the tree, suggesting that T107A might have arisen from multiple distinct base substitution events. Shannon entropy analysis showed high variability in certain capsid sites, but none overlapped with known lenacapavir resistance sites.
Conclusions: These findings suggest a low prevalence of naturally occurring lenacapavir resistance mutations in Uganda, supporting lenacapavir's potential efficacy in this region.
© The Author(s) 2025. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures

References
-
- Gilead . Sunlenca® (lenacapavir) Receives FDA Approval as a First-in-Class, Twice-Yearly Treatment Option for People Living with Multi-Drug Resistant HIV. 2022. https://www.gilead.com/news-and-press/press-room/press-releases/2022/12/...
-
- FDA . FDA Approves New HIV Drug for Adults With Limited Treatment Options. 2022. https://www.hiv.gov/blog/fda-approves-new-hiv-drug-for-adults-with-limit...
-
- Gilead Sciences, Inc . Gilead Announces First Global Regulatory Approval of Sunlenca® (Lenacapavir), The Only Twice-Yearly HIV Treatment Option. 2022. https://www.gilead.com/news-and-press/press-room/press-releases/2022/8/g...
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

