Systematic empirical evaluation of individual base editing targets: Validating therapeutic targets in USH2A and comparison of methods
- PMID: 39881543
- PMCID: PMC11997516
- DOI: 10.1016/j.ymthe.2025.01.042
Systematic empirical evaluation of individual base editing targets: Validating therapeutic targets in USH2A and comparison of methods
Abstract
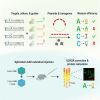
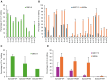
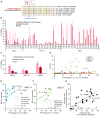
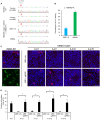
Base editing shows promise for the correction of human mutations at a higher efficiency than other repair methods and is especially attractive for mutations in large genes that are not amenable to gene augmentation therapy. Here, we demonstrate a comprehensive workflow for in vitro screening of potential therapeutic base editing targets for the USH2A gene and empirically validate the efficiency of adenine and cytosine base editor/guide combinations for correcting 35 USH2A mutations. Editing efficiency and bystander edits are compared between different target templates (plasmids vs. transgenes) and assays (next-generation sequencing vs. Sanger), as well as comparisons between unbiased empirical results and computational predictions. Based on these observations, practical assay recommendations are discussed. Finally, a humanized knockin mouse model was created with the best-performing target, the nonsense mutation c.11864G>A p.(Trp3955∗). Split-intein AAV9 delivery of editing reagents resulted in the restoration of USH2A protein and a correction rate of 65% ± 3% at the mutant base pair and of 52% ± 3% excluding bystander amino acid changes. This efficiency is higher than that seen in a retinal gene editing program testing in a clinical trial. These results demonstrate the effectiveness of this overall strategy to identify and test base editing reagents with the potential for human therapeutic applications.
Keywords: AAV; USH2A; Usher syndrome; adenine base editor; base editing; cytosine base editor; photoreceptors; retina; retinitis pigmentosa.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Patent US20230159913A1 was issued based on the reported work. J.C. has received consulting payments from Applied Genetic Technologies Corporation, Beam Therapeutics, Biogen, Gensight Biologics, Octant Bio, Wave Life Sciences, and Vedere. Q.L. has received consulting payments from Editas Medicine and Entrada Therapeutics. Y.T. received salary support from his employer, Daiichi Sankyo Co., Ltd., while acting as a visiting scientist at Mass Eye and Ear. D.R.L. is a consultant and equity owner of Beam Therapeutics, Prime Medicine, Pairwise Plants, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or genome engineering agents. J.M.L. is currently an employee of Prime Medicine.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

