A yeast-based reverse genetics system to generate HCoV-OC43 reporter viruses encoding an eighth subgenomic RNA
- PMID: 39882907
- PMCID: PMC11852775
- DOI: 10.1128/jvi.01671-24
A yeast-based reverse genetics system to generate HCoV-OC43 reporter viruses encoding an eighth subgenomic RNA
Abstract
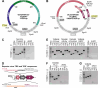
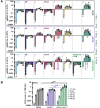
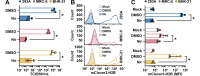
Coronaviruses have large, positive-sense single-stranded RNA genomes that challenge conventional strategies for mutagenesis. Yeast genetics has been used to manipulate large viral genomes, including those of herpesviruses and coronaviruses. This method, known as transformation-associated recombination (TAR), involves assembling complete viral genomes from dsDNA copies of viral genome fragments via homologous recombination in Saccharomyces cerevisiae. Here, we report our development of a TAR assembly and mutagenesis system for the endemic, seasonal human coronavirus (HCoV) strain OC43. HCoV-OC43 generally causes mild respiratory symptoms and is classified as a biosafety level 2 agent, making it useful for studying fundamental aspects of coronavirus biology and for comparative studies of more highly pathogenic betacoronaviruses. Following cDNA synthesis from HCoV-OC43 viral RNA, we generated five plasmids encompassing ~7.2 kb portions of the ORF1ab gene, the NS2 to M segment, or the N gene and structured to facilitate reporter gene insertions in the M-to-N intergenic region. Using these plasmids, we completed independent assemblies of yeast centromeric plasmids encoding ORF1ab, NS2a to N, as well as full-length HCoV-OC43 plasmids. A wild-type virus (OC43YA), as well as mClover3-H2B (OC43-mCloYA), mRuby3-H2B (OC43-mRubyYA), and mCardinal (OC43-mCardYA) reporter viruses, were rescued. The OC43-mCloYA reporter virus replicated comparably to an OC43 reference strain and produced the mClover3-H2B protein from a novel subgenomic RNA through insertion of an eighth body transcription regulatory sequence, preventing the need to delete or mutate viral genes. This updated HCoV-OC43 reverse genetics system will contribute to a better understanding of betacoronavirus host-pathogen interactions and can accelerate studies of novel antivirals.
Importance: Coronaviruses are ubiquitous pathogens that infect humans resulting in both mild and severe respiratory infections. Human coronavirus strain OC43 (HCoV-OC43) is one of many viruses responsible for common colds and is a useful model of more severe coronavirus infections. In this study, we describe an updated HCoV-OC43 mutagenesis system that uses yeast to capture six DNA fragments of the viral RNA genome and assemble them into full-length genomes in yeast/bacterial plasmids. The design of this system allowed for the rapid assembly and rescue of functional HCoV-OC43 viruses, including fluorescent reporter viruses with expanded genetic capacity. This updated reverse genetics system will enhance our ability to monitor viral replication, through building new reporter viruses, while also enhancing the study of betacoronavirus biology through the generation of mutant HCoV-OC43 viruses.
Keywords: HCoV-OC43; TAR; coronavirus; mutagenesis; reporter virus; reverse genetics; transformation-associated recombination; virus; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

