Acetylation modification in the regulation of macroautophagy
- PMID: 39883319
- PMCID: PMC11740868
- DOI: 10.1007/s44307-024-00027-7
Acetylation modification in the regulation of macroautophagy
Abstract
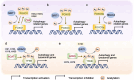
Macroautophagy, commonly referred to as autophagy, is an evolutionarily conserved cellular process that plays a crucial role in maintaining cellular homeostasis. It orchestrates the delivery of dysfunctional or surplus cellular materials to the vacuole or lysosome for degradation and recycling, particularly during adverse conditions. Over the past few decades, research has unveiled intricate regulatory mechanisms governing autophagy through various post-translational modifications (PTMs). Among these PTMs, acetylation modification has emerged as a focal point in yeast and animal studies. It plays a pivotal role in autophagy by directly targeting core components within the central machinery of autophagy, including autophagy initiation, nucleation, phagophore expansion, and autophagosome maturation. Additionally, acetylation modulates autophagy at the transcriptional level by modifying histones and transcription factors. Despite its well-established significance in yeast and mammals, the role of acetylation in plant autophagy remains largely unexplored, and the precise regulatory mechanisms remain enigmatic. In this comprehensive review, we summarize the current understanding of the function and underlying mechanisms of acetylation in regulating autophagy across yeast, mammals, and plants. We particularly highlight recent advances in deciphering the impact of acetylation on plant autophagy. These insights not only provide valuable guidance but also inspire further scientific inquiries into the intricate role of acetylation in plant autophagy.
Keywords: Acetylation; Autophagy; Deacetylation; Lysine; Post-translational modification.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All authors approved the final manuscript and the submission to this journal. Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- Annunziata I, van de Vlekkert D, Wolf E, Finkelstein D, Neale G, Machado E, Mosca R, Campos Y, Tillman H, Roussel MF, Andrew Weesner J, Ellen Fremuth L, Qiu X, Han M, Grosveld GC, D Azzo A. MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat Commun. 2019;10:3623. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

