Importance of pre-mRNA splicing and its study tools in plants
- PMID: 39883322
- PMCID: PMC11740881
- DOI: 10.1007/s44307-024-00009-9
Importance of pre-mRNA splicing and its study tools in plants
Abstract
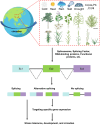
Alternative splicing (AS) significantly enriches the diversity of transcriptomes and proteomes, playing a pivotal role in the physiology and development of eukaryotic organisms. With the continuous advancement of high-throughput sequencing technologies, an increasing number of novel transcript isoforms, along with factors related to splicing and their associated functions, are being unveiled. In this review, we succinctly summarize and compare the different splicing mechanisms across prokaryotes and eukaryotes. Furthermore, we provide an extensive overview of the recent progress in various studies on AS covering different developmental stages in diverse plant species and in response to various abiotic stresses. Additionally, we discuss modern techniques for studying the functions and quantification of AS transcripts, as well as their protein products. By integrating genetic studies, quantitative methods, and high-throughput omics techniques, we can discover novel transcript isoforms and functional splicing factors, thereby enhancing our understanding of the roles of various splicing modes in different plant species.
Keywords: Alternative splicing; Modern biotechnology; Plant development; Splicing machinery; Stress response.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors have no competing interests to declare that are relevant to the content of this article.
Figures

References
-
- Bao YJ, Chen JX, Zhang YJ, Fernie AR, Zhang J, Huang BX, Zhu FY, Cao FL. Emerging role of jasmonic acid in woody plant development. Advanced Agrochem. 2023; ISSN 2773-2371.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

