Mechanism of microbial action of the inoculated nitrogen-fixing bacterium for growth promotion and yield enhancement in rice (Oryza sativa L.)
- PMID: 39883349
- PMCID: PMC11709144
- DOI: 10.1007/s44307-024-00038-4
Mechanism of microbial action of the inoculated nitrogen-fixing bacterium for growth promotion and yield enhancement in rice (Oryza sativa L.)
Abstract
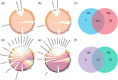
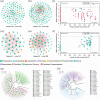
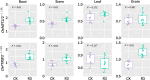
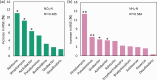
The use of nitrogen-fixing bacteria in agriculture is increasingly recognized as a sustainable method to boost crop yields, reduce chemical fertilizer use, and improve soil health. However, the microbial mechanisms by which inoculation with nitrogen-fixing bacteria enhance rice production remain unclear. In this study, rice seedlings were inoculated with the nitrogen-fixing bacterium R3 (Herbaspirillum) at the rhizosphere during the seedling stage in a pot experiment using paddy soil. We investigated the effects of such inoculation on nutrient content in the rhizosphere soil, plant growth, and the nitrogen-fixing microbial communities within the rhizosphere and endorhizosphere. The findings showed that inoculation with the R3 strain considerably increased the amounts of nitrate nitrogen, ammonium nitrogen, and available phosphorus in the rhizosphere by 14.77%, 27.83%, and 22.67%, respectively, in comparison to the control (CK). Additionally, the theoretical yield of rice was enhanced by 8.81% due to this inoculation, primarily through a 10.24% increase in the effective number of rice panicles and a 4.14% increase in the seed setting rate. Further analysis revealed that the structure of the native nitrogen-fixing microbial communities within the rhizosphere and endorhizosphere were altered by inoculation with the R3 strain, significantly increasing the α-diversity of the communities. The relative abundance of key nitrogen-fixing genera such as Ralstonia, Azotobacter, Geobacter, Streptomyces, and Pseudomonas were increased, enhancing the quantity and community stability of the nitrogen-fixing community. Consequently, the nitrogen-fixing capacity and sustained activity of the microbial community in the rhizosphere soil were strengthened. Additionally, the expression levels of the nitrogen absorption and transport-related genes OsNRT1 and OsPTR9 in rice roots were upregulated by inoculation with the R3 strain, potentially contributing to the increased rice yield. Our study has revealed the potential microbial mechanisms through which inoculation with nitrogen-fixing bacteria enhances rice yield. This finding provides a scientific basis for subsequent agricultural practices and is of critical importance for increasing rice production and enhancing the ecosystem services of rice fields.
Keywords: nifH gene; Keystone taxa; Microbiome; Nitrogen-fixing bacterium; Rice yield; α-diversity.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All authors agreed with the content and that all gave explicit consent to submit. Competing interests: Author J.L. is a member of the Editorial Board for Advanced Biotechnology, but she is not involved in the journal’s review of, and decisions related to, this manuscript.
Figures


References
-
- Chaia EE, Wall LG, Huss-Danell K. Life in soil by the actinorhizal root nodule endophyte Frankia. A Review Symbiosis. 2010;51:201–26. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

