Therapeutic gene correction of HBB frameshift CD41-42 (-TCTT) deletion in human hematopoietic stem cells
- PMID: 39883359
- PMCID: PMC11740860
- DOI: 10.1007/s44307-024-00053-5
Therapeutic gene correction of HBB frameshift CD41-42 (-TCTT) deletion in human hematopoietic stem cells
Abstract
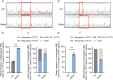
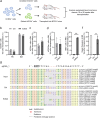
Β-thalassemia is one of the global health burdens. The CD41-42 (-TCTT) mutation at HBB is the most prevalent pathogenic mutation of β-thalassemia in both China and Southeast Asia. Previous studies focused on repairing the HBB CD41-42 (-TCTT) mutation in β-thalassemia patient-specific induced pluripotent stem cells, which were subsequently differentiated into hematopoietic stem and progenitor cells (HSPCs) for transplantation. In this study, we directly applied the CRISPR/Cas9-based gene editing therapy to correct the HBB CD41-42 (-TCTT) mutation in patient-derived HSPCs. The effective editing induced by Cas9:sgRNA ribonucleoprotein and single-stranded oligodeoxynucleotides (ssODNs) was confirmed in HUDEP-2 cell lines harboring the HBB CD41-42 (-TCTT) mutation. Further correction of heterozygote and homozygote HBB CD41-42 (-TCTT) mutations in patient-derived HSPCs resulted in a 13.4-40.8% increase in the proportion of HBB-expressing (HBB +) cells following erythroid differentiation in vitro. At 16 weeks post-xenotransplantation of the edited HSPCs into coisogenic immunodeficient mice, the reparation efficiency in engrafted bone marrow was 17.21% ± 3.66%. Multiparameter flow cytometric analysis of the engrafted bone marrow showed an increase in the percentage of HBB + cells without impairing the ability of engraftment, self-renewal, and multilineage hematopoietic repopulation of HSPCs. For the safety evaluation, 103 potential off-target sites were predicted by SITE-seq and CRISPOR, with one site displaying significant off-target editing. Since this off-target site is located in the intergenic region, it is presumed to pose minimal risk. Taken together, our study provides critical preclinical data supporting the safety and efficacy of the gene therapy approach for HBB CD41-42 (-TCTT) mutation.
Keywords: HBB CD41-42 (-TCTT); CRISPR/Cas9; Gene editing therapy; Hematopoietic stem and progenitor cells; β-thalassemia.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The human CD34 + HSPCs were obtained from mobilized peripheral blood of patients with HBB CD41-42 (-TCTT) mutation through apheresis at Sun Yat-sen Memorial Hospital, with approval from the Institutions’ Ethical Committee (No. 2020–328). All patients provided informed consent. All the animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (SYSU-IACUC-2020-B079) and followed the legal requirements in China and Guangdong Province. Competing interests: Hui Xu, Ying Luo, Lin Cheng, and Junbin Liang are employees of Reforgene Medicine. Author Junjiu Huang is a member of the Editorial Board for Advanced Biotechnology, and the author is not involved in the journal’s review of and decisions related to this manuscript.
Figures



References
-
- Andreani, M., Testi, M., Gaziev, J., Condello, R., Bontadini, A., Tazzari, P. L., . . . Lucarelli, G. (2011). Quantitatively different red cell/nucleated cell chimerism in patients with long-term, persistent hematopoietic mixed chimerism after bone marrow transplantation for thalassemia major or sickle cell disease. Haematologica, 96(1), 128–133. 10.3324/haematol.2010.031013 - PMC - PubMed
-
- Cameron, P., Fuller, C. K., Donohoue, P. D., Jones, B. N., Thompson, M. S., Carter, M. M., . . . May, A. P. (2017). Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods, 14(6), 600–606. 10.1038/nmeth.4284 - PubMed
LinkOut - more resources
Full Text Sources

