Elevated protein lactylation promotes immunosuppressive microenvironment and therapeutic resistance in pancreatic ductal adenocarcinoma
- PMID: 39883522
- PMCID: PMC11957693
- DOI: 10.1172/JCI187024
Elevated protein lactylation promotes immunosuppressive microenvironment and therapeutic resistance in pancreatic ductal adenocarcinoma
Abstract
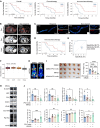
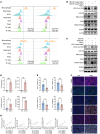
Metabolic reprogramming shapes the tumor microenvironment (TME) and may lead to immunotherapy resistance in pancreatic ductal adenocarcinoma (PDAC). Elucidating the impact of pancreatic cancer cell metabolism in the TME is essential to therapeutic interventions. "Immune cold" PDAC is characterized by elevated lactate levels resulting from tumor cell metabolism, abundance of protumor macrophages, and reduced cytotoxic T cells in the TME. Analysis of fluorine-18 fluorodeoxyglucose (18F-FDG) uptake in patients showed that increased global protein lactylation in PDAC correlates with worse clinical outcomes in immunotherapy. Inhibition of lactate production in pancreatic tumors via glycolysis or mutant-KRAS inhibition reshaped the TME, thereby increasing their sensitivity to immune checkpoint blockade (ICB) therapy. In pancreatic tumor cells, lactate induces K63 lactylation of endosulfine α (ENSA-K63la), a crucial step that triggers STAT3/CCL2 signaling. Consequently, elevated CCL2 secreted by tumor cells facilitates tumor-associated macrophage (TAM) recruitment to the TME. High levels of lactate also drive transcriptional reprogramming in TAMs via ENSA-STAT3 signaling, promoting an immunosuppressive environment. Targeting ENSA-K63la or CCL2 enhances the efficacy of ICB therapy in murine and humanized pancreatic tumor models. In conclusion, elevated lactylation reshapes the TME and promotes immunotherapy resistance in PDAC. A therapeutic approach targeting ENSA-K63la or CCL2 has shown promise in sensitizing pancreatic cancer immunotherapy.
Keywords: Cancer; Cancer immunotherapy; Immunology; Macrophages; Oncology.
Figures





Comment in
- Targeting lactylation and the STAT3/CCL2 axis to overcome immunotherapy resistance in pancreatic ductal adenocarcinoma doi: 10.1172/JCI191422
References
-
- Hu ZI, O’Reilly EM. Therapeutic developments in pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2024;21(1):7–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

