Comprehensive analysis of ceRNA Networks in UCEC: Prognostic and therapeutic implications
- PMID: 39883704
- PMCID: PMC11781699
- DOI: 10.1371/journal.pone.0314314
Comprehensive analysis of ceRNA Networks in UCEC: Prognostic and therapeutic implications
Abstract
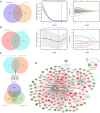
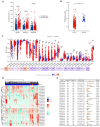
Endometrial cancer (UCEC) is the most prevalent gynecological malignancy in high-income countries, and its incidence is rising globally. Although early-stage UCEC can be treated with surgery, advanced cases have a poor prognosis, highlighting the need for effective molecular biomarkers to improve diagnosis and prognosis. In this study, we analyzed mRNA and miRNA sequencing data from UCEC tissues and adjacent non-cancerous tissues from the TCGA database. Differential expression analysis was conducted using the DESeq2 package, identifying differentially expressed lncRNAs, miRNAs, and mRNAs (DElncRNAs, DEmiRNAs, and DEmRNAs). Key molecules were screened using LASSO regression, and a ceRNA network was constructed by predicting lncRNA-miRNA and miRNA-mRNA interaction, which were visualized with Cytoscape. Functional enrichment analysis elucidated the roles and mechanisms of the network. The prognostic potential of the identified RNAs was assessed through survival and Cox regression analyses, while methylation and immune infiltration analyses explored regulatory mechanisms and immune interactions. We identified a prognostic lncRNA-miRNA-mRNA ceRNA network in UCEC, centered on the CDKN2B-AS1-hsa-miR-497-5p-IGF2BP3 axis. Survival analyses confirmed the prognostic significance of this network, with univariate Cox regression demonstrating a strong association between its aberrant expression and overall prognosis in UCEC. However, multivariate Cox regression suggested that other clinical factors may modulate this relationship. Methylation analysis revealed low methylation levels of IGF2BP3, possibly contributing to its overexpression. Furthermore, immune infiltration studies highlighted significant correlations between CDKN2B-AS1, IGF2BP3, and multiple immune cell types, suggesting that this axis regulates the tumor immune microenvironment. These findings suggest that the CDKN2B-AS1-hsa-miR-497-5p-IGF2BP3 axis is a key regulatory element in UCEC and a potential therapeutic target.
Copyright: © 2025 Fan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

