Uterine organoids reveal insights into epithelial specification and plasticity in development and disease
- PMID: 39883834
- PMCID: PMC11804710
- DOI: 10.1073/pnas.2422694122
Uterine organoids reveal insights into epithelial specification and plasticity in development and disease
Abstract
Understanding how epithelial cells in the female reproductive tract (FRT) differentiate is crucial for reproductive health, yet the underlying mechanisms remain poorly defined. At birth, FRT epithelium is highly malleable, allowing differentiation into various epithelial types, but the regulatory pathways guiding these early cell fate decisions are unclear. Here, we use neonatal mouse endometrial organoids and assembloid coculture models to investigate how innate cellular plasticity and external mesenchymal signals influence epithelial differentiation. Our findings demonstrate that uterine epithelium undergoes marked age-dependent changes, transitioning from a highly plastic state capable of forming both monolayered and multilayered structures to a more restricted fate as development progresses. Interestingly, parallels emerge between the developmental plasticity of neonatal uterine epithelium and pathological conditions such as endometrial cancer, where similar regulatory mechanisms may reactivate, driving abnormal epithelial differentiation and tumorigenesis. These results not only deepen our understanding of early uterine development but also offer a valuable model for studying the progression of reproductive diseases and cancers.
Keywords: assembloids; development; epithelium; organoid; uterus.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

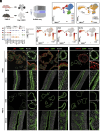
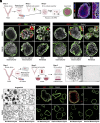


References
-
- Kurita T., Cunha G. R., Robboy S. J., Mills A. A., Medina R. T., Differential expression of p63 isoforms in female reproductive organs. Mech. Dev. 122, 1043–1055 (2005). - PubMed
MeSH terms
Grants and funding
- 1R37HD114609/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R37 HD114609/HD/NICHD NIH HHS/United States
- R01 HD112315/HD/NICHD NIH HHS/United States
- MU/Paula and Rodger Riney Foundation
- UL1TR002389-07/HHS | NIH | National Center for Advancing Translational Sciences (NCATS)
LinkOut - more resources
Full Text Sources

