Upregulated FoxO1 promotes arrhythmogenesis in mice with heart failure and preserved ejection fraction
- PMID: 39885127
- PMCID: PMC11782541
- DOI: 10.1038/s41467-025-56186-1
Upregulated FoxO1 promotes arrhythmogenesis in mice with heart failure and preserved ejection fraction
Abstract
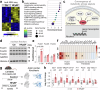
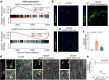
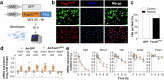
Myocardial fibrosis leads to cardiac dysfunction and arrhythmias in heart failure with preserved ejection fraction (HFpEF), but the underlying mechanisms remain poorly understood. Here, RNA sequencing identifies Forkhead Box1 (FoxO1) signaling as abnormal in male HFpEF hearts. Genetic suppression of FoxO1 alters the intercellular communication between cardiomyocytes and fibroblasts, alleviates abnormal diastolic relaxation, and reduces arrhythmias. Targeted downregulation of FoxO1 in activated fibroblasts reduces cardiac fibrosis, blunts arrhythmogenesis and improves diastolic function in HFpEF. These results not only implicate FoxO1 in arrhythmogenesis and lusitropy but also demonstrate that pro-fibrotic remodeling and cardiomyocyte-fibroblast communication can be corrected, constituting an alternative therapeutic strategy for HFpEF.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Redfield, M. M. & Borlaug, B. A. Heart failure with preserved ejection fraction: a review. JAMA329, 827–838 (2023). - PubMed
MeSH terms
Substances
Grants and funding
- CLIN1-14874/California Institute for Regenerative Medicine (CIRM)
- R01 HL155346/HL/NHLBI NIH HHS/United States
- EDUC4-12751/California Institute for Regenerative Medicine (CIRM)
- 24IPA1275047/American Heart Association (American Heart Association, Inc.)
- DISC2-13009/California Institute for Regenerative Medicine (CIRM)
- PR150620/United States Department of Defense | United States Army | Army Medical Command | Congressionally Directed Medical Research Programs (CDMRP)
- 940033/American Heart Association (American Heart Association, Inc.)
- R01 HL135866/HL/NHLBI NIH HHS/United States
- R01 HL147570/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

