Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
- PMID: 39885130
- PMCID: PMC11782662
- DOI: 10.1038/s41467-025-56468-8
Phase separation of a microtubule plus-end tracking protein into a fluid fractal network
Abstract
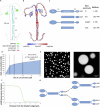
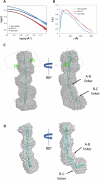
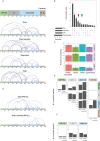
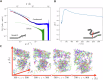
Microtubule plus-end tracking proteins (+TIPs) participate in nearly all microtubule-based cellular processes and have recently been proposed to function as liquid condensates. However, their formation and internal organization remain poorly understood. Here, we have study the phase separation of Bik1, a CLIP-170 family member and key +TIP involved in budding yeast cell division. Bik1 is a dimer with a rod-shaped conformation primarily defined by its central coiled-coil domain. Its liquid condensation likely involves the formation of higher-order oligomers that phase separate in a manner dependent on the protein's N-terminal CAP-Gly domain and C-terminal EEY/F-like motif. This process is accompanied by conformational rearrangements in Bik1, leading to at least a two-fold increase in multivalent interactions between its folded and disordered domains. Unlike classical liquids, Bik1 condensates exhibit a heterogeneous, fractal supramolecular structure with protein- and solvent-rich regions. This structural evidence supports recent percolation-based models of biomolecular condensates. Together, our findings offer insights into the structure, dynamic rearrangement, and organization of a complex, oligomeric, and multidomain protein in both dilute and condensed states. Our experimental framework can be applied to other biomolecular condensates, including more complex +TIP networks.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Akhmanova, A. & Steinmetz, M. O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol.16, 711–726 (2015). - PubMed
-
- Akhmanova, A. & Hoogenraad, C. C. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol.17, 47–54 (2005). - PubMed
MeSH terms
Substances
Grants and funding
- CRSII5_189940/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- 884104/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 871037/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Miscellaneous

