Advancing the genetic engineering toolbox by combining AsCas12a knock-in mice with ultra-compact screening
- PMID: 39885149
- PMCID: PMC11782673
- DOI: 10.1038/s41467-025-56282-2
Advancing the genetic engineering toolbox by combining AsCas12a knock-in mice with ultra-compact screening
Abstract
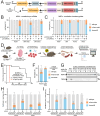
Cas12a is a next-generation gene editing tool that enables multiplexed gene targeting. Here, we present a mouse model that constitutively expresses enhanced Acidaminococcus sp. Cas12a (enAsCas12a) linked to an mCherry fluorescent reporter. We demonstrate efficient single and multiplexed gene editing in vitro, using primary and transformed cells from enAsCas12a mice. We further demonstrate successful in vivo gene editing, using normal and cancer-prone enAsCas12a stem cells to reconstitute the haematopoietic system of wild-type mice. We also present compact, genome-wide Cas12a knockout libraries, with four crRNAs per gene encoded across one (Scherzo) or two (Menuetto) vectors, and demonstrate the utility of these libraries across methodologies: in vitro enrichment and drop-out screening in lymphoma cells and immortalised fibroblasts, respectively, and in vivo screens to identify lymphoma-driving events. Finally, we demonstrate CRISPR multiplexing via simultaneous gene knockout (via Cas12a) and activation (via dCas9-SAM) using primary T cells and fibroblasts. Our enAsCas12a mouse and accompanying crRNA libraries enhance genome engineering capabilities and complement current CRISPR technologies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.M.D., L.H., and J.P.F. are current employees of Genentech, and B.H. and V.S. have previously been employees of Genentech, where the Cas12a mouse libraries presented in this work were developed. The remaining authors declare no competing interests.
Figures


References
-
- Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature532, 517–521 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

