Identification and validation of a prognostic signature of drug resistance and mitochondrial energy metabolism-related differentially expressed genes for breast cancer
- PMID: 39885513
- PMCID: PMC11780791
- DOI: 10.1186/s12967-025-06080-7
Identification and validation of a prognostic signature of drug resistance and mitochondrial energy metabolism-related differentially expressed genes for breast cancer
Abstract
Background: Drug resistance constitutes one of the principal causes of poor prognosis in breast cancer patients. Although cancer cells can maintain viability independently of mitochondrial energy metabolism, they remain reliant on mitochondrial functions for the synthesis of new DNA strands. This dependency underscores a potential link between mitochondrial energy metabolism and drug resistance. Hence, drug resistance and mitochondrial energy metabolism-related differentially expressed genes (DMRDEGs) may emerge as candidates for novel cancer biomarkers. This study endeavors to assess the viability of DMRDEGs as biomarkers or therapeutic targets for breast cancer.
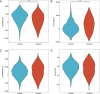
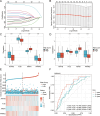
Methods: We utilized the DRESIS database and MSigDB to identify genes related to drug resistance. Additionally, we sourced genes associated with mitochondrial energy metabolism from GeneCards and extant literature. By merging these genes with differentially expressed genes observed in normal and tumor tissues from the TCGA-BRCA and GEO databases, we successfully identified the DMRDEGs. Employing unsupervised consensus clustering, we divided breast cancer patients into two distinct groups based on the DMRDEGs. Consequently, we identified four hub genes to formulate a prognostic model, applying Cox regression, LASSO regression, and Random Forest methods. Furthermore, we examined immune infiltration and tumor mutation burden of the genes within our model and scrutinized divergences in the immune microenvironment between high- and low-risk groups. Small hairpin RNA and lentiviral plasmids were designed for stable transfection of breast cancer cell lines MDA-MB-231 and HCC1806. By conducting clone formation, scratch test, transwell assays, cell viability assay and measurement of oxygen consumption we initiated a preliminary investigation into mechanistic roles of AIFM1.
Results: We utilized DMRDEGs to develop a prognostic model that includes four mRNAs for breast cancer. This model combined with various clinical features and critical breast cancer facets, demonstrated remarkable efficacy in predicting patient outcomes. AIFM1 appeared to enhance the proliferation, migration, and invasiveness of breast cancer cell lines MDA-MB-231 and HCC1806. Moreover, by reducing oxygen consumption, it aids in the cancer cells' acquisition of drug resistance.
Conclusions: DMRDEGs hold promise as diagnostic markers and therapeutic targets for breast cancer. Among the associated mutated genes, ATP7B, FUS, AIFM1, and PPARG could serve as early diagnostic indicators, and notably, AIFM1 may present itself as a promising therapeutic target.
Keywords: Breast cancer; Drug resistance; Mitochondrial energy metabolism; Prognostic model.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: All authors confirm their consent for publication the manuscript. Competing interests: The authors declare that there is no conflict of interest.
Figures













References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. GLOBOCAN estimates of incidence and mor tality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2022. 10.3322/caac.21834.
-
- Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397:1750–69. - PubMed
-
- Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F. Breast cancer. Nat Rev Dis Primers. 2019;5:66. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

