Classification performance and reproducibility of GPT-4 omni for information extraction from veterinary electronic health records
- PMID: 39885843
- PMCID: PMC11780673
- DOI: 10.3389/fvets.2024.1490030
Classification performance and reproducibility of GPT-4 omni for information extraction from veterinary electronic health records
Abstract
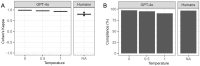
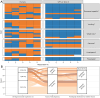
Large language models (LLMs) can extract information from veterinary electronic health records (EHRs), but performance differences between models, the effect of hyperparameter settings, and the influence of text ambiguity have not been previously evaluated. This study addresses these gaps by comparing the performance of GPT-4 omni (GPT-4o) and GPT-3.5 Turbo under different conditions and by investigating the relationship between human interobserver agreement and LLM errors. The LLMs and five humans were tasked with identifying six clinical signs associated with feline chronic enteropathy in 250 EHRs from a veterinary referral hospital. When compared to the majority opinion of human respondents, GPT-4o demonstrated 96.9% sensitivity [interquartile range (IQR) 92.9-99.3%], 97.6% specificity (IQR 96.5-98.5%), 80.7% positive predictive value (IQR 70.8-84.6%), 99.5% negative predictive value (IQR 99.0-99.9%), 84.4% F1 score (IQR 77.3-90.4%), and 96.3% balanced accuracy (IQR 95.0-97.9%). The performance of GPT-4o was significantly better than that of its predecessor, GPT-3.5 Turbo, particularly with respect to sensitivity where GPT-3.5 Turbo only achieved 81.7% (IQR 78.9-84.8%). GPT-4o demonstrated greater reproducibility than human pairs, with an average Cohen's kappa of 0.98 (IQR 0.98-0.99) compared to 0.80 (IQR 0.78-0.81) with humans. Most GPT-4o errors occurred in instances where humans disagreed [35/43 errors (81.4%)], suggesting that these errors were more likely caused by ambiguity of the EHR than explicit model faults. Using GPT-4o to automate information extraction from veterinary EHRs is a viable alternative to manual extraction, but requires validation for the intended setting to ensure accuracy and reliability.
Keywords: Chat-GPT; Real-World Data (RWD); Real-World Evidence (RWE); artificial intelligence; feline chronic enteropathy; generative-pretrained transformers; machine learning; text mining.
Copyright © 2025 Wulcan, Jacques, Lee, Kovacs, Dausend, Prince, Wulcan, Marsilio and Keller.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Center for Veterinary Medicine. CVM GFI #266 Use of Real-World Data and Real-World Evidence to Support Effectiveness of New Animal Drugs. FDA: (2021). Available at: https://www.fda.gov/regulatory-information/search-fda-guidance-documents... (accessed December 17, 2024).
-
- Brown TB, Mann B, Ryder N, Subbiah M, Kaplan J, Dhariwal P, et al. . Language models are few-shot learners. arXiv [preprint]. (2020). 10.48550/arXiv.2005.14165 - DOI

