Consistent features observed in structural probing data of eukaryotic RNAs
- PMID: 39885881
- PMCID: PMC11780854
- DOI: 10.1093/nargab/lqaf001
Consistent features observed in structural probing data of eukaryotic RNAs
Abstract
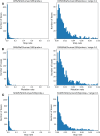
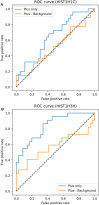
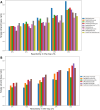
Understanding RNA structure is crucial for elucidating its regulatory mechanisms. With the recent commercialization of messenger RNA vaccines, the profound impact of RNA structure on stability and translation efficiency has become increasingly evident, underscoring the importance of understanding RNA structure. Chemical probing of RNA has emerged as a powerful technique for investigating RNA structure in living cells. This approach utilizes chemical probes that selectively react with accessible regions of RNA, and by measuring reactivity, the openness and potential of RNA for protein binding or base pairing can be inferred. Extensive experimental data generated using RNA chemical probing have significantly contributed to our understanding of RNA structure in cells. However, it is crucial to acknowledge potential biases in chemical probing data to ensure an accurate interpretation. In this study, we comprehensively analyzed transcriptome-scale RNA chemical probing data in eukaryotes and report common features. Notably, in all experiments, the number of bases modified in probing was small, the bases showing the top 10% reactivity well reflected the known secondary structure, bases with high reactivity were more likely to be exposed to solvent and low reactivity did not reflect solvent exposure, which is important information for the analysis of RNA chemical probing data.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures



Similar articles
-
Single-Molecule Correlated Chemical Probing: A Revolution in RNA Structure Analysis.Acc Chem Res. 2023 Apr 4;56(7):763-775. doi: 10.1021/acs.accounts.2c00782. Epub 2023 Mar 14. Acc Chem Res. 2023. PMID: 36917683 Free PMC article.
-
Carbodiimide reagents for the chemical probing of RNA structure in cells.RNA. 2019 Jan;25(1):135-146. doi: 10.1261/rna.067561.118. Epub 2018 Nov 2. RNA. 2019. PMID: 30389828 Free PMC article.
-
Probing-directed identification of novel structured RNAs.RNA Biol. 2016;13(2):232-42. doi: 10.1080/15476286.2015.1132140. RNA Biol. 2016. PMID: 26732206 Free PMC article.
-
The role of RNA structure in 3' end processing in eukaryotes.Curr Opin Struct Biol. 2024 Dec;89:102933. doi: 10.1016/j.sbi.2024.102933. Epub 2024 Sep 29. Curr Opin Struct Biol. 2024. PMID: 39348742 Review.
-
SHAPE Directed Discovery of New Functions in Large RNAs.Acc Chem Res. 2021 May 18;54(10):2502-2517. doi: 10.1021/acs.accounts.1c00118. Epub 2021 May 7. Acc Chem Res. 2021. PMID: 33960770 Free PMC article. Review.
References
-
- Cech T.R., Steitz J.A. The noncoding RNA revolution—trashing old rules to forge new ones. Cell. 2014; 157:77–94. - PubMed
-
- Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982; 31:147–157. - PubMed
-
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. - PubMed
-
- Metkar M., Pepin C.S., Moore M.J. Tailor made: the art of therapeutic mRNA design. Nat. Rev. Drug Discov. 2024; 23:67–83. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

