Role of hepatocyte RIPK1 in maintaining liver homeostasis during metabolic challenges
- PMID: 39886919
- PMCID: PMC11785375
- DOI: 10.7554/eLife.96798
Role of hepatocyte RIPK1 in maintaining liver homeostasis during metabolic challenges
Abstract
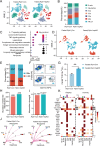
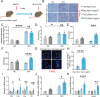
As a central hub for metabolism, the liver exhibits strong adaptability to maintain homeostasis in response to food fluctuations throughout evolution. However, the mechanisms governing this resilience remain incompletely understood. In this study, we identified Receptor interacting protein kinase 1 (RIPK1) in hepatocytes as a critical regulator in preserving hepatic homeostasis during metabolic challenges, such as short-term fasting or high-fat dieting. Our results demonstrated that hepatocyte-specific deficiency of RIPK1 sensitized the liver to short-term fasting-induced liver injury and hepatocyte apoptosis in both male and female mice. Despite being a common physiological stressor that typically does not induce liver inflammation, short-term fasting triggered hepatic inflammation and compensatory proliferation in hepatocyte-specific RIPK1-deficient (Ripk1-hepKO) mice. Transcriptomic analysis revealed that short-term fasting oriented the hepatic microenvironment into an inflammatory state in Ripk1-hepKO mice, with up-regulated expression of inflammation and immune cell recruitment-associated genes. Single-cell RNA sequencing further confirmed the altered cellular composition in the liver of Ripk1-hepKO mice during fasting, highlighting the increased recruitment of macrophages to the liver. Mechanically, our results indicated that ER stress was involved in fasting-induced liver injury in Ripk1-hepKO mice. Overall, our findings revealed the role of RIPK1 in maintaining liver homeostasis during metabolic fluctuations and shed light on the intricate interplay between cell death, inflammation, and metabolism.
Keywords: ER stress; RIPK1; fasting; immunology; inflammation; liver homeostasis; mouse.
© 2024, Zhang et al.
Conflict of interest statement
WZ, HL, DZ, YY, LT, YZ, SH, XZ, QS, PL, YW, WL, JZ, HZ, YC, DW No competing interests declared
Figures





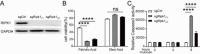
Update of
- doi: 10.1101/2024.03.04.583302
- doi: 10.7554/eLife.96798.1
- doi: 10.7554/eLife.96798.2
References
-
- Bernardo-Seisdedos G, Bilbao J, Fernández-Ramos D, Lopitz-Otsoa F, Gutierrez de Juan V, Bizkarguenaga M, Mateos B, Fondevila MF, Abril-Fornaguera J, Diercks T, Lu SC, Nogueiras R, Mato JM, Millet O. Metabolic landscape of the mouse liver by quantitative31 p nuclear magnetic resonance analysis of the phosphorome. Hepatology. 2021;74:148–163. doi: 10.1002/hep.31676. - DOI - PMC - PubMed
-
- Bettermann K, Vucur M, Haybaeck J, Koppe C, Janssen J, Heymann F, Weber A, Weiskirchen R, Liedtke C, Gassler N, Müller M, de Vos R, Wolf MJ, Boege Y, Seleznik GM, Zeller N, Erny D, Fuchs T, Zoller S, Cairo S, Buendia MA, Prinz M, Akira S, Tacke F, Heikenwalder M, Trautwein C, Luedde T. TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell. 2010;17:481–496. doi: 10.1016/j.ccr.2010.03.021. - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

