Beyond the mono-nucleosome
- PMID: 39887339
- PMCID: PMC12224902
- DOI: 10.1042/BST20230721
Beyond the mono-nucleosome
Abstract
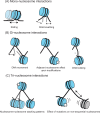
Nucleosomes, the building block of chromatin, are responsible for regulating access to the DNA sequence. This control is critical for essential cellular processes, including transcription and DNA replication and repair. Studying chromatin can be challenging both in vitro and in vivo, leading many to use a mono-nucleosome system to answer fundamental questions relating to chromatin regulators and binding partners. However, the mono-nucleosome fails to capture essential features of the chromatin structure, such as higher-order chromatin folding, local nucleosome-nucleosome interactions, and linker DNA trajectory and flexibility. We briefly review significant discoveries enabled by the mono-nucleosome and emphasize the need to go beyond this model system in vitro. Di-, tri-, and tetra-nucleosome arrays can answer important questions about chromatin folding, function, and dynamics. These multi-nucleosome arrays have highlighted the effects of varying linker DNA lengths, binding partners, and histone post-translational modifications in a more chromatin-like environment. We identify various chromatin regulatory mechanisms yet to be explored with multi-nucleosome arrays. Combined with in-solution biophysical techniques, studies of minimal multi-nucleosome chromatin models are feasible.
Keywords: chromatin; chromatin remodeler; multi-nucleosome array; nucleosome.
© 2025 The Author(s); published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

