A cellular model of TDP-43 induces phosphorylated TDP-43 aggregation with distinct changes in solubility and autophagy dysregulation
- PMID: 39887552
- PMCID: PMC12310990
- DOI: 10.1111/febs.17413
A cellular model of TDP-43 induces phosphorylated TDP-43 aggregation with distinct changes in solubility and autophagy dysregulation
Abstract
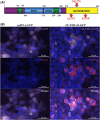
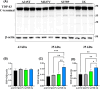
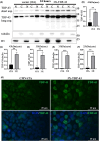
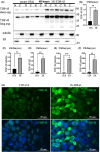
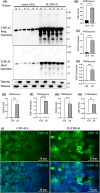
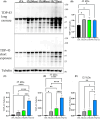
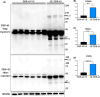
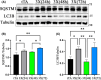
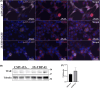
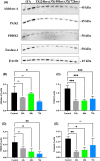
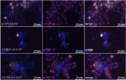
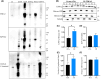
Amyotrophic lateral sclerosis (ALS) is an incurable neurodegenerative disease that affects neurons in the brain and spinal cord, causing loss of muscle control, and eventually leads to death. Phosphorylated transactive response DNA binding protein-43 (TDP-43) is the major pathological protein in both sporadic and familial ALS, forming cytoplasmic aggregates in over 95% of cases. Of the 10-15% of ALS cases that are familial, mutations in TDP-43 represent about 5% of those with a family history. We have developed an in vitro overexpression model by introducing three familial ALS mutations (A315T, M337V, and S379P) in the TDP-43 (TARDBP) gene which we define as 3X-TDP-43. This overexpression model TDP-43 shows deficits in autophagy flux and colocalization of TDP-43 with stress granules. We also observe a progressive shift of TDP-43 to the cytoplasm in this model. This overexpression model shows a reduction in solubility of phosphorylated TDP-43 from RIPA to urea soluble. Four glycolytic enzymes, phosphoglycerate kinase one (PGK1), aldolase A (ALDOA), enolase 1 (ENO1), and pyruvate dehydrogenase kinase 1 (PDK1) show significant time-dependent decreases in 3X-TDP-43 expressing cells. Shotgun proteomic analysis shows global changes in the importin subunit alpha-1 (KPNA2), heat shock 70 kDa protein 1A (HSPA1A), and protein disulfide-isomerase A3 (PDIA3) expression levels and coimmunoprecipitation reveals that these proteins complex with TDP-43. Overall, these results suggest that the 3X-TDP-43 model may provide new insights into pathophysiology and an avenue for drug screening in vitro for those suffering from ALS and related TDP-43 proteinopathies.
Keywords: ALS; TDP‐43; aggregation; autophagy; glycolysis; stress granules.
© 2025 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Wang HY, Wang IF, Bose J & Shen CK (2004) Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83, 130–139. - PubMed
-
- Buratti E, De Conti L, Stuani C, Romano M, Baralle M & Baralle F (2010) Nuclear factor TDP‐43 can affect selected microRNA levels. FEBS J 277, 2268–2281. - PubMed
-
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra‐Lantz C & Shoesmith C (2007) TDP43 is a human low molecular weight neurofilament (hNFL) mRNA‐binding protein. Mol Cell Neurosci 35, 320–327. - PubMed
MeSH terms
Substances
Grants and funding
- 52008702/HHMI
- T32 GM144895/GM/NIGMS NIH HHS/United States
- 2023004/National Science Foundation
- P30 GM145765/GM/NIGMS NIH HHS/United States
- P20GM103446/GM/NIGMS NIH HHS/United States
- P20 GM103446/GM/NIGMS NIH HHS/United States
- 0823/The Paul H. Boerger Fund of the Delaware Community Foundation
- P20 GM103653/GM/NIGMS NIH HHS/United States
- R25 GM122722/GM/NIGMS NIH HHS/United States
- P20GM103653/GM/NIGMS NIH HHS/United States
- T32GM144895-03/GM/NIGMS NIH HHS/United States
- P20GM103446/GM/NIGMS NIH HHS/United States
- P20GM103653/GM/NIGMS NIH HHS/United States
- T32GM144895-03/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

