Combining genome-wide association and genomic prediction to unravel the genetic architecture of carotenoid accumulation in carrot
- PMID: 39887573
- PMCID: PMC11782711
- DOI: 10.1002/tpg2.20560
Combining genome-wide association and genomic prediction to unravel the genetic architecture of carotenoid accumulation in carrot
Abstract
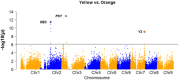
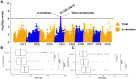
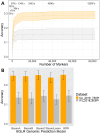
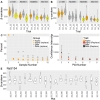
Carrots (Daucus carota L.) are a rich source of provitamin A, namely, α- and β-carotene. Breeding programs prioritize increasing β-carotene content for improved color and nutrition. Understanding the genetic basis of carotenoid accumulation is crucial for implementing genomic-assisted selection to develop high-carotenoid lines. While previous studies identified loci (Y2, Y, Or, and REC) associated with carrot color and carotenoid content, this study employed genome-wide association (GWA) in a diverse panel of 738 carrot accessions. We discovered a novel locus with a candidate gene encoding phytoene synthase, a key enzyme in carotenoid biosynthesis. The Y2, Y, Or, and REC loci are mostly fixed in orange varieties, yet considerable variation in carotenoid concentration persists. This suggests a multigenic trait influenced by the environment. GWA of carotenoid concentration identified a quantitative trait locus for total carotenoids and α-carotene. We explored the accuracy of genomic prediction (GP) models to predict carotenoid concentration. We determined the optimal number of plants and plots required for accurate carotenoid phenotyping, finding ≥5 plants per plot and three plots per site as the minimum effective sample per accession. GP models achieved accuracies ranging from 0.06 to 0.40 depending on the carotenoid measured and environment the carrots were assayed. Additional studies in breeding programs will clarify the potential of genomic-assisted selection for high-carotenoid carrots.
© 2025 The Author(s). The Plant Genome published by Wiley Periodicals LLC on behalf of Crop Science Society of America.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Banga, O. (1957). Effect of some environmental factors on the carotene content of carrots. IVT, 92, 796–805. - PubMed
-
- Banga, O. , & De Bruyn, J. W. (1956). Selection of carrots for carotene content: III Planting distances and ripening equilibrium of the roots. Euphytica, 5, 87–93. 10.1007/BF00021287 - DOI
-
- Bernardo, R. (2008). Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Science, 48, 1649–1664. 10.2135/cropsci2008.03.0131 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

