Effect of Notch1 signaling on muscle engraftment and maturation from pluripotent stem cells
- PMID: 39889709
- PMCID: PMC11864150
- DOI: 10.1016/j.stemcr.2024.102396
Effect of Notch1 signaling on muscle engraftment and maturation from pluripotent stem cells
Abstract
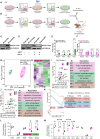
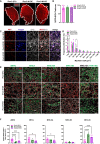
Pax3-induced pluripotent stem cell-derived myogenic progenitors display an embryonic molecular signature but become postnatal upon transplantation. Because this correlates with upregulation of Notch signaling, here we probed whether NOTCH1 is required for in vivo maturation by performing gain- and loss-of-function studies in inducible Pax3 (iPax3) myogenic progenitors. Transplantation studies revealed that Notch1 signaling did not change the number of donor-derived fibers; however, the NOTCH1 overexpression cohorts showed enhanced satellite cell engraftment and more mature fibers, as indicated by fewer fibers expressing the embryonic myosin heavy-chain isoform and more type IIX fibers. While donor-derived Pax7+ cells were detected in all transplants, in the absence of Notch1, secondary grafts exhibited a high fraction of these cells in the interstitial space, indicating that NOTCH1 is required for proper satellite cell homing. Transcriptional profiling of NOTCH1-modified donor-derived satellite cells suggests that this may be due to changes in the extracellular matrix organization, cell cycle, and metabolism.
Keywords: NOTCH1; Pax3; fiber type; muscle regeneration; myogenic progenitors; pluripotent stem cells; satellite cells; secondary transplantation.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.C.R.P. is the cofounder and holds equity in Myogenica.
Figures


References
-
- Azzag K., Ortiz-Cordero C., Oliveira N.A.J., Magli A., Selvaraj S., Tungtur S., Upchurch W., Iaizzo P.A., Lu Q.L., Perlingeiro R.C.R. Efficient engraftment of pluripotent stem cell-derived myogenic progenitors in a novel immunodeficient mouse model of limb girdle muscular dystrophy 2I. Skelet. Muscle. 2020;10:10. doi: 10.1186/s13395-020-00228-3. - DOI - PMC - PubMed
-
- Baik J., Ortiz-Cordero C., Magli A., Azzag K., Crist S.B., Yamashita A., Kiley J., Selvaraj S., Mondragon-Gonzalez R., Perrin E., et al. Establishment of Skeletal Myogenic Progenitors from Non-Human Primate Induced Pluripotent Stem Cells. Cells. 2023;12 doi: 10.3390/cells12081147. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

