NSm is a critical determinant for bunyavirus transmission between vertebrate and mosquito hosts
- PMID: 39890788
- PMCID: PMC11785797
- DOI: 10.1038/s41467-024-54809-7
NSm is a critical determinant for bunyavirus transmission between vertebrate and mosquito hosts
Abstract
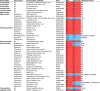
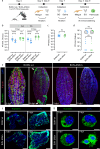
Bunyavirales is a very large order including viruses infecting a variety of taxonomic groups such as arthropods, vertebrates, plants, and protozoa. Some bunyaviruses are transmitted between vertebrate hosts by blood-sucking arthropods and cause major diseases in humans and animals. It is not understood why only some bunyaviruses have evolved the capacity to be transmitted by arthropod vectors. Here we show that only vector-borne bunyaviruses express a non-structural protein, NSm, whose function has so far remained largely elusive. Using as experimental system Bunyamwera virus (BUNV) and its invertebrate host, Aedes aegypti, we show that NSm is dispensable for viral replication in mosquito cells in vitro but is absolutely required for successful infection in the female mosquito following a blood meal. More specifically, NSm is required for cell-to-cell spread and egress from the mosquito midgut, a known barrier to viral infection. Notably, the requirement for NSm is specific to the midgut; bypassing this barrier by experimental intrathoracic infection of the mosquito eliminates the necessity of NSm for virus spread in other tissues, including the salivary glands. Overall, we unveiled a key evolutionary process that allows the transmission of vector-borne bunyaviruses between arthropod and vertebrate hosts.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Tajudeen, Y. A. et al. Emerging Arboviruses of Public Health Concern in Africa: Priorities for Future Research and Control Strategies. Challenges13, 60 (2022). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

