Cellular senescence-associated gene IFI16 promotes HMOX1-dependent evasion of ferroptosis and radioresistance in glioblastoma
- PMID: 39890789
- PMCID: PMC11785807
- DOI: 10.1038/s41467-025-56456-y
Cellular senescence-associated gene IFI16 promotes HMOX1-dependent evasion of ferroptosis and radioresistance in glioblastoma
Abstract
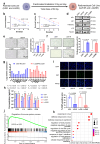
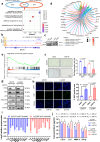
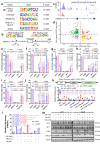
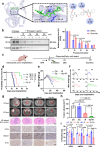
Glioblastoma multiforme (GBM) remains a therapeutic challenge due to its aggressive nature and recurrence. This study establishes a radioresistant GBM cell model through repeated irradiation and observes a cellular senescence-like phenotype in these cells. Comprehensive genomic and transcriptomic analyses identify IFI16 as a central regulator of this phenotype and contributes to radioresistance. IFI16 activates HMOX1 transcription thereby attenuating ferroptosis by reducing lipid peroxidation, ROS production, and intracellular Fe2+ content following irradiation. Furthermore, IFI16 interacts with the transcription factors JUND and SP1 through its pyrin domain, robustly facilitating HMOX1 expression, further inhibiting ferroptosis and enhancing radioresistance in GBM. Notably, glyburide, a sulfonylurea compound, effectively disrupts IFI16 function and enhances ferroptosis and radiosensitivity. By targeting the pyrin domain of IFI16, glyburide emerges as a potential therapeutic agent against GBM radioresistance. These findings underscore the central role of IFI16 in GBM radioresistance and offer promising avenues to improve GBM treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

