Use of brain MRI and gene expression atlases to reconstruct the pathophysiology of autoimmune neurological disorders: The proof-of-concept of NMOSD
- PMID: 39891565
- PMCID: PMC11789429
- DOI: 10.1177/13524585241307154
Use of brain MRI and gene expression atlases to reconstruct the pathophysiology of autoimmune neurological disorders: The proof-of-concept of NMOSD
Abstract
Background: The understanding of disease pathophysiology is pivotal for tailored treatments. The spatial distribution of brain damage relies on the regional antigen expression and the local balance of susceptibility and protective elements.
Objective: As proof-of-concept, we investigated the spatial association between brain damage and gene expression in aquaporin-4-IgG-positive neuromyelitis optica spectrum disorder (AQP4 + NMOSD).
Methods: In this multicenter cross-sectional study, 90 AQP4 + NMOSD patients and 94 age-matched healthy controls underwent a brain magnetic resonance imaging (MRI). We used T2-hyperintense lesion probability maps and white/gray matter atrophy as proxies of inflammation and neurodegeneration. The association with the expression of 266 candidate genes was obtained with the Multimodal Environment for Neuroimaging and Genomic Analysis platform. A functional-enrichment analysis investigated overrepresented biological processes.
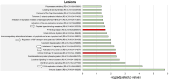
Results: In AQP4 + NMOSD, T2-hyperintense lesions were mainly periventricular; atrophy mostly involved the visual pathway. The expression of AQP4 and complement (C4a and C5) was associated with both inflammation and neurodegeneration. Complement activation and regulation/uptake of the insulin-like growth factor were the most relevant enriched pathways. Nonspecific pathways related to DNA synthesis and repair were associated with brain atrophy.
Conclusions: Quantitative MRI and gene expression atlas identified the key elements of AQP4 + NMOSD pathophysiology. This analysis could help in understanding the pathophysiology of antibody-mediated autoimmune disorders.
Keywords: Neuromyelitis optica spectrum disorders; gene expression; magnetic resonance imaging.
Conflict of interest statement
Declaration of Conflicting InterestsThe author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: L.C. received speaker and consultant honoraria from ACCMED, Roche, BMS Celgene, and Sanofi. L.S. received grants and contracts from FISM—Fondazione Italiana Sclerosi Multipla—within a fellowship program (cod. 2019/BR/009) and received speakers’ honoraria from Biogen. E.P. received speaker honoraria from Biogen Idec. P.P. received speaker honoraria from Roche, Biogen, Novartis, Merck Serono, Bristol-Myers Squibb, Genzyme, Horizon, and Sanofi; he received research support from the Italian Ministry of Health and Fondazione Italiana Sclerosi Multipla. S.M. has been an advisor or speaker for Medis, Merck, Teva, Hemofarm, and Roche. V.M. received honoraria for consulting services or speaking activity from Biogen, Merck, Novartis, TEVA, Almirall, and Sanofi. L.M. received compensation for speaking activities, and/or consulting services from Merck, Biogen, Novartis, Roche, Sanofi, and TEVA. M.R. reports no disclosures. J.I. reports no disclosures. O.T. has been a speaker for Medis, Merck, Teva, Hemofarm, Novartis, and Roche. J.D. has been an advisor or speaker for Bayer HealthCare, Sanofi-Genzyme, Medis, Merck, Teva, Novartis, Biogen, Hemofarm, and Roche. M.F. is Editor-in-Chief of the
Figures




References
-
- Yamamura T, Kleiter I, Fujihara K, et al.. Trial of satralizumab in neuromyelitis optica spectrum disorder. N Engl J Med 2019; 381: 2114–2124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

