Defining hypoxia in cancer: A landmark evaluation of hypoxia gene expression signatures
- PMID: 39892389
- PMCID: PMC11872601
- DOI: 10.1016/j.xgen.2025.100764
Defining hypoxia in cancer: A landmark evaluation of hypoxia gene expression signatures
Abstract
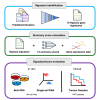
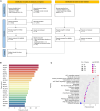
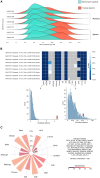
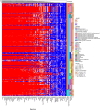
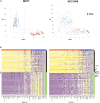
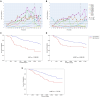
Tumor hypoxia drives metabolic shifts, cancer progression, and therapeutic resistance. Challenges in quantifying hypoxia have hindered the exploitation of this potential "Achilles' heel." While gene expression signatures have shown promise as surrogate measures of hypoxia, signature usage is heterogeneous and debated. Here, we present a systematic pan-cancer evaluation of 70 hypoxia signatures and 14 summary scores in 104 cell lines and 5,407 tumor samples using 472 million length-matched random gene signatures. Signature and score choice strongly influenced the prediction of hypoxia in vitro and in vivo. In cell lines, the Tardon signature was highly accurate in both bulk and single-cell data (94% accuracy, interquartile mean). In tumors, the Buffa and Ragnum signatures demonstrated superior performance, with Buffa/mean and Ragnum/interquartile mean emerging as the most promising for prospective clinical trials. This work delivers recommendations for experimental hypoxia detection and patient stratification for hypoxia-targeting therapies, alongside a generalizable framework for signature evaluation.
Keywords: biomarkers; gene signature; hypoxia; hypoxia-targeting therapies; patient stratification; radiotherapy; signature scores; single cell; transcriptomics; tumorigenesis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
-
- Harris A.L. Hypoxia — a key regulatory factor in tumour growth. Nat. Rev. Cancer. 2002;2:38–47. - PubMed
-
- Brizel D.M., Dodge R.K., Clough R.W., Dewhirst M.W. Oxygenation of head and neck cancer: changes during radiotherapy and impact on treatment outcome. Radiother. Oncol. 1999;53:113–117. - PubMed
-
- Nordsmark M., Overgaard J. Tumor hypoxia is independent of hemoglobin and prognostic for loco-regional tumor control after primary radiotherapy in advanced head and neck cancer. Acta Oncol. 2004;43:396–403. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

