ATRX mutations mediate an immunogenic phenotype and macrophage infiltration in neuroblastoma
- PMID: 39892705
- PMCID: PMC12057689
- DOI: 10.1016/j.canlet.2025.217495
ATRX mutations mediate an immunogenic phenotype and macrophage infiltration in neuroblastoma
Abstract
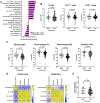
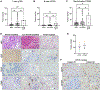
ATRX is one of the most frequently mutated genes in high-risk neuroblastoma. ATRX mutations are mutually exclusive with MYCN amplification and mark a recognizable patient subgroup, presenting in older children with chemotherapy-resistant, slowly progressive disease. The mechanisms underlying how ATRX mutations drive high-risk and difficult-to-treat neuroblastoma are still largely elusive. To unravel the role of ATRX in neuroblastoma, we generated isogenic neuroblastoma cell line models with ATRX loss-of-function and ATRX in-frame multi-exon deletions, representing different types of alterations found in patients. RNA-sequencing analysis consistently showed significant upregulation of inflammatory response pathways in the ATRX-altered cell lines. In vivo, ATRX alterations are consistently associated with macrophage infiltration across multiple xenograft models. Furthermore, ATRX alterations also result in upregulation of epithelial-to-mesenchymal transition pathways and a reduction in expression of adrenergic core-regulatory circuit genes. Consistent with this, bioinformatic analysis of previously published neuroblastoma patient data sets revealed that ATRX-altered neuroblastomas display an immunogenic phenotype and higher score of macrophages (with no distinction between M1 and M2 macrophage populations) and dendritic cells, but not lymphocytes. Histopathological assessment of diagnostic samples from patients with ATRX mutant disease confirmed these findings with significantly more macrophage infiltration compared to MYCN-amplified tumors. In conclusion, we show that gene expression and cell-state changes as a result of ATRX alterations associate with a characteristic immune cell infiltration in both in vivo models and patient samples. Together, this provides novel insight into mechanisms underlying the distinct clinical phenotype seen in this group of patients.
Keywords: ATRX; Macrophages; Neuroblastoma; Tumour microenvironment.
Copyright © 2025. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Aguilera P, López-Contreras AJ, ATRX, a guardian of chromatin, Trends Genet. 39 (6) (2023) 505–519. - PubMed
-
- Jansky S, Sharma AK, Körber V, Quintero A, Toprak UH, Wecht EM, et al. , Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma, Nat. Genet 53 (5) (2021) 683–693. - PubMed
-
- Qiu B, Matthay KK, Advancing therapy for neuroblastoma, Nat. Rev. Clin. Oncol 19 (8) (2022) 515–533. - PubMed
-
- Morgenstern DA, Pötschger U, Moreno L, Papadakis V, Owens C, Ash S, et al. , Risk stratification of high-risk metastatic neuroblastoma: a report from the HRNBL-1/SIOPEN study, Pediatr. Blood Cancer 65 (11) (2018) e27363. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

