Cellular Crosstalk Promotes Hepatic Progenitor Cell Proliferation and Stellate Cell Activation in 3D Co-culture
- PMID: 39892785
- PMCID: PMC11968293
- DOI: 10.1016/j.jcmgh.2025.101472
Cellular Crosstalk Promotes Hepatic Progenitor Cell Proliferation and Stellate Cell Activation in 3D Co-culture
Abstract
Background & aims: Following liver damage, ductular reaction often coincides with liver fibrosis. Proliferation of hepatic progenitor cells is observed in ductular reaction, whereas activated hepatic stellate cells (HSCs) are the main drivers of liver fibrosis. These observations may suggest a functional interaction between these 2 cell types. Here, we report on an in vitro co-culture system to examine these interactions and validate their co-expression in human liver explants.
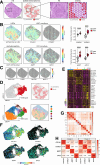
Methods: In a 3D organoid co-culture system, we combined freshly isolated quiescent mouse HSCs and fluorescently labeled progenitor cells (undifferentiated intrahepatic cholangiocyte organoids), permitting real-time observation of cell morphology and behavior. After 7 days, cells were sorted based on the fluorescent label and analyzed for changes in gene expression.
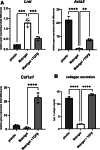
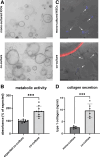
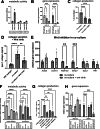
Results: In the 3D co-culture system, the proliferation of progenitor cells is enhanced, and HSCs are activated, recapitulating the cellular events observed in the patient liver. Both effects in 3D co-culture require close contact between the 2 different cell types. HSC activation during 3D co-culture differs from quiescent (3D mono-cultured) HSCs and activated HSCs on plastic (2D mono-culture). Upregulation of a cluster of genes containing Aldh1a2, Cthrc1, and several genes related to frizzled binding/Wnt signaling were exclusively observed in 3D co-cultured HSCs. The localized co-expression of specific genes was confirmed by spatial transcriptomics in human liver explants.
Conclusion: An in vitro 3D co-culture system provides evidence for direct interactions between HSCs and progenitor cells, which are sufficient to drive responses that are similar to those seen during ductular reaction and fibrosis. This model paves the way for further research into the cellular basis of liver pathology.
Keywords: Ductular Reaction; Intrahepatic Cholangiocyte Organoid; Liver Fibrosis; Retinoic Acid.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Asrani S.K., Devarbhavi H., Eaton J., Kamath P.S. Burden of liver diseases in the world. J Hepatol. 2019;70:151–171. - PubMed
-
- Rinella M.E., Lazarus J.V., Ratziu V., et al. NAFLD Nomenclature consensus group A multi-society Delphi consensus statement on new fatty liver disease nomenclature. Ann Hepatol. 2024;29 - PubMed
-
- Roskams T.A., Theise N.D., Balabaud C., et al. Nomenclature of the finer branches of the biliary tree: canals, ductules, and ductular reactions in human livers. Hepatology. 2004;39:1739–1745. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

