Deep-ER: Deep Learning ECCENTRIC Reconstruction for fast high-resolution neurometabolic imaging
- PMID: 39894238
- PMCID: PMC11952141
- DOI: 10.1016/j.neuroimage.2025.121045
Deep-ER: Deep Learning ECCENTRIC Reconstruction for fast high-resolution neurometabolic imaging
Abstract
Introduction: Altered neurometabolism is an important pathological mechanism in many neurological diseases and brain cancer, which can be mapped non-invasively by Magnetic Resonance Spectroscopic Imaging (MRSI). Advanced MRSI using non-cartesian compressed-sense acquisition enables fast high-resolution metabolic imaging but has lengthy reconstruction times that limits throughput and needs expert user interaction. Here, we present a robust and efficient Deep Learning reconstruction embedded in a physical model within an end-to-end automated processing pipeline to obtain high-quality metabolic maps.
Methods: Fast high-resolution whole-brain metabolic imaging was performed at 3.4 mm3 isotropic resolution with acquisition times between 4:11-9:21 min:s using ECCENTRIC pulse sequence on a 7T MRI scanner. Data were acquired in a high-resolution phantom and 27 human participants, including 22 healthy volunteers and 5 glioma patients. A deep neural network using recurring interlaced convolutional layers with joint dual-space feature representation was developed for deep learning ECCENTRIC reconstruction (Deep-ER). 21 subjects were used for training and 6 subjects for testing. Deep-ER performance was compared to iterative compressed sensing Total Generalized Variation reconstruction using image and spectral quality metrics.
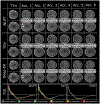
Results: Deep-ER demonstrated 600-fold faster reconstruction than conventional methods, providing improved spatial-spectral quality and metabolite quantification with 12%-45% (P<0.05) higher signal-to-noise and 8%-50% (P<0.05) smaller Cramer-Rao lower bounds. Metabolic images clearly visualize glioma tumor heterogeneity and boundary. Deep-ER generalizes reliably to unseen data.
Conclusion: Deep-ER provides efficient and robust reconstruction for sparse-sampled MRSI. The accelerated acquisition-reconstruction MRSI is compatible with high-throughput imaging workflow. It is expected that such improved performance will facilitate basic and clinical MRSI applications for neuroscience and precision medicine.
Keywords: Brain; Compressed sensing; Deep learning; Glioma; Image reconstruction; MR spectroscopic imaging; Metabolism; Non-cartesian; Ultra high field.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



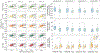
References
-
- Bagchi Sonali, Mitra Sanjit K., 1996. The nonuniform discrete Fourier transform and its applications in filter design. I. 1-D. IEEE Trans. Circuits Syst. II 43 (6), 422–433.
-
- Bogner Wolfgang, Gruber Stephan, Trattnig Siegfried, Chmelik Marek, 2012. High-resolution mapping of human brain metabolites by free induction decay 1H MRSI at 7 T. NMR Biomed. 25 (6), 873–882. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

