Translational regulation of SND1 governs endothelial homeostasis during stress
- PMID: 39895626
- PMCID: PMC11785924
- DOI: 10.1172/JCI168730
Translational regulation of SND1 governs endothelial homeostasis during stress
Abstract
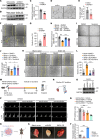
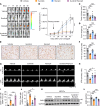
Translational control shapes the proteome and is particularly important in regulating gene expression under stress. A key source of endothelial stress is treatment with tyrosine kinase inhibitors (TKIs), which lowers cancer mortality but increases cardiovascular mortality. Using a human induced pluripotent stem cell-derived endothelial cell (hiPSC-EC) model of sunitinib-induced vascular dysfunction combined with ribosome profiling, we assessed the role of translational control in hiPSC-ECs in response to stress. We identified staphylococcal nuclease and tudor domain-containing protein 1 (SND1) as a sunitinib-dependent translationally repressed gene. SND1 translational repression was mediated by the mTORC1/4E-BP1 pathway. SND1 inhibition led to endothelial dysfunction, whereas SND1 OE protected against sunitinib-induced endothelial dysfunction. Mechanistically, SND1 transcriptionally regulated UBE2N, an E2-conjugating enzyme that mediates K63-linked ubiquitination. UBE2N along with the E3 ligases RNF8 and RNF168 regulated the DNA damage repair response pathway to mitigate the deleterious effects of sunitinib. In silico analysis of FDA-approved drugs led to the identification of an ACE inhibitor, ramipril, that protected against sunitinib-induced vascular dysfunction in vitro and in vivo, all while preserving the efficacy of cancer therapy. Our study established a central role for translational control of SND1 in sunitinib-induced endothelial dysfunction that could potentially be therapeutically targeted to reduce sunitinib-induced vascular toxicity.
Keywords: Cancer; Cardiovascular disease; Endothelial cells; Vascular biology.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

