HMGA1 acts as an epigenetic gatekeeper of ASCL2 and Wnt signaling during colon tumorigenesis
- PMID: 39895630
- PMCID: PMC11785931
- DOI: 10.1172/JCI184442
HMGA1 acts as an epigenetic gatekeeper of ASCL2 and Wnt signaling during colon tumorigenesis
Abstract
Mutated tumor cells undergo changes in chromatin accessibility and gene expression, resulting in aberrant proliferation and differentiation, although how this occurs is unclear. HMGA1 chromatin regulators are abundant in stem cells and oncogenic in diverse tissues; however, their role in colon tumorigenesis is only beginning to emerge. Here, we uncover a previously unknown epigenetic program whereby HMGA1 amplifies Wnt signaling during colon tumorigenesis driven by inflammatory microbiota and/or Adenomatous polyposis coli (Apc) inactivation. Mechanistically, HMGA1 "opens" chromatin to upregulate the stem cell regulator, Ascl2, and downstream Wnt effectors, promoting stem and Paneth-like cell states while depleting differentiated enterocytes. Loss of just one Hmga1 allele within colon epithelium restrains tumorigenesis and Wnt signaling driven by mutant Apc and inflammatory microbiota. However, HMGA1 deficiency has minimal effects in colon epithelium under homeostatic conditions. In human colon cancer cells, HMGA1 directly induces ASCL2 by recruiting activating histone marks. Silencing HMGA1 disrupts oncogenic properties, whereas reexpression of ASCL2 partially rescues these phenotypes. Further, HMGA1 and ASCL2 are coexpressed and upregulated in human colorectal cancer. Together, our results establish HMGA1 as an epigenetic gatekeeper of Wnt signals and cell state under conditions of APC inactivation, illuminating HMGA1 as a potential therapeutic target in colon cancer.
Keywords: Colorectal cancer; Epigenetics; Oncology; Transcription.
Conflict of interest statement
Figures





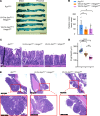



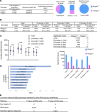


Comment in
- HMGA1 is a crucial mediator of colon tumorigenesis driven by the loss of APC doi: 10.1172/JCI187442
References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

