EFHD1 Activates SIK3 to Limit Colorectal Cancer Initiation and Progression via the Hippo Pathway
- PMID: 39895792
- PMCID: PMC11786025
- DOI: 10.7150/jca.103229
EFHD1 Activates SIK3 to Limit Colorectal Cancer Initiation and Progression via the Hippo Pathway
Abstract
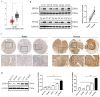
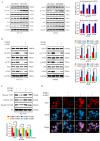
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers, with high rates of metastasis and lethality. EF-hand domain-containing protein D1 (EFHD1) and salt-inducible kinase 3 (SIK3) have been studied in several cancer types. Aberrant expression of EFHD1 and SIK3 has been observed in CRC, but little research has addressed their regulatory abilities and signaling pathways. In this study, we aimed to explore the efficacy of EFHD1 in inhibiting CRC proliferation and metastasis and to elucidate the underlying mechanisms involved in the upregulation of SIK3 expression. Cell viability, colony formation, wound healing, Transwell assay, orthotopic xenograft, and pulmonary metastasis mouse models were used to detect the antiproliferative and anti-metastatic effects of EFHD1 against CRC in vitro and in vivo. The Gene Expression Profiling Interactive Analysis (GEPIA) database was used to determine EFHD1 and SIK3 expression in CRC. The regulatory roles of EFHD1 and SIK3 in mediating anti-metastatic effects in CRC were measured using western blotting, immunohistochemical, and immunofluorescence analyses. The results showed that EFHD1 expression was significantly repressed in the clinical CRC samples. EFHD1 markedly suppressed cell proliferation, migration, and invasion in vitro and inhibited tumor growth and metastasis in vivo. Analysis of the GEPIA database revealed that EFHD1 expression positively correlated with SIK3 expression. SIK3 overexpression inhibited the migration of CRC cells, and SIK3 knockdown partially eliminated the inhibitory effects of EFHD1 on CRC metastasis. EFHD1 exerted anti-metastatic effects against CRC via upregulating SIK3 and inhibiting epithelial-mesenchymal transition (EMT) processing through modulating the Hippo signaling pathway. Collectively, these findings identify EFHD1 as a potent SIK3 agonist and highlight the EFHD1-SIK3 axis as a key modulator of the Hippo signaling pathway in CRC. EFHD1 serves as a novel regulator and is worthy of further development as a novel therapeutic target in CRC.
Keywords: Colorectal cancer; EFHD1; EMT; Hippo pathway; SIK3.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Niehues JM, Quirke P, West NP, Grabsch HI, van Treeck M, Schirris Y, Veldhuizen GP, Hutchins G, Richman SD, Foersch S, Brinker TJ, Fukuoka J, Bychkov A, Uegami W, Truhn D, Brenner H, Brobeil A, Hoffmeister M, Kather JN. Generalizable biomarker prediction from cancer pathology slides with self-supervised deep learning: a retrospective multi-centric study. Cell Rep Med. 2023;4:100980. - PMC - PubMed
-
- Baviskar T, Momin M, Liu J, Guo B, Bhatt L. Target Genetic Abnormalities for the Treatment of Colon Cancer and its Progression to Metastasis. Curr Drug Targets. 2021;22:722–733. - PubMed
LinkOut - more resources
Full Text Sources

