Deep plasma and tissue proteome profiling of knockout mice reveals pathways associated with Svep1 deficiency
- PMID: 39895831
- PMCID: PMC11782998
- DOI: 10.1016/j.jmccpl.2025.100283
Deep plasma and tissue proteome profiling of knockout mice reveals pathways associated with Svep1 deficiency
Abstract
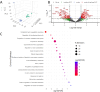
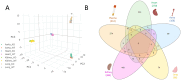
Despite strong causal associations with cardiovascular and metabolic disorders including coronary artery disease, hypertension, and type 2 diabetes, as well as a range of other diseases, the exact function of the protein SVEP1 remains largely unknown. Animal models have been employed to investigate how SVEP1 contributes to disease, with a focus on murine models exploring its role in development, cardiometabolic disease and platelet biology. In this study, we aimed to comprehensively phenotype the proteome of Svep1 +/- mice compared to wild-type (WT) littermates using liquid chromatography-tandem mass spectrometry (LC-MS/MS) bottom-up proteomics in plasma, heart, aorta, lung, and kidney to identify dysregulated pathways and biological functions associated with Svep1 deficiency. Our findings reveal that Svep1 deficiency leads to significant proteomic alterations across the mouse, with the highest number of dysregulated proteins observed in plasma and kidney. Key dysregulated proteins in plasma include upregulation of ADGRV1, CDH1, and MYH6, and downregulation of MTIF2 and AKAP13 which, alongside other proteins dysregulated across tissues, indicate disruption in cell adhesion, extracellular matrix organisation, platelet degranulation, and Rho GTPase pathways. Novel findings include significant enrichment of complement cascades in plasma, suggesting dysregulation of innate immune responses and hemostasis due to Svep1 deficiency. Pathways related to chylomicron assembly and lipid metabolism were also enriched. Additionally, we developed a high-throughput quantitative targeted LC-MS/MS assay to measure endogenous levels of murine SVEP1. SVEP1 was detectable in lung homogenate and showed a significant reduction in SVEP1 levels in Svep1 +/- vs. WT, but was not identified in plasma, heart, aorta, or kidney, likely due to expression levels below the assay's detection limit. Overall, this deep phenotyping study provides insight into the systemic impact of Svep1 deficiency.
Keywords: Cardiovascular disease; Deep phenotyping; Extracellular matrix; Knockout mouse; Liquid chromatography-mass spectrometry; Proteomics; SVEP1.
© 2025 The Author(s).
Conflict of interest statement
The authors declare they have no competing interests.
Figures







References
-
- Shur I., Socher R., Hameiri M., Fried A., Benayahu D. Molecular and cellular characterization of SEL-OB/SVEP1 in osteogenic cells in vivo and in vitro. J Cell Physiol. 2006;206(2):420–427. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

