Characterization of three novel Helicobacter species infecting stomachs of dogs and cats: Helicobacter gastrocanis sp. nov., Helicobacter gastrofelis sp. nov., and Helicobacter felistomachi sp. nov
- PMID: 39895936
- PMCID: PMC11783220
- DOI: 10.3389/fmicb.2024.1459401
Characterization of three novel Helicobacter species infecting stomachs of dogs and cats: Helicobacter gastrocanis sp. nov., Helicobacter gastrofelis sp. nov., and Helicobacter felistomachi sp. nov
Abstract
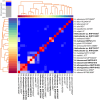
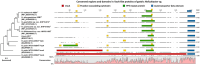
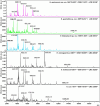
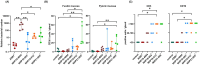
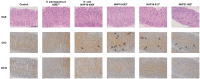
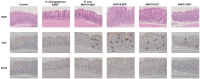
Helicobacter species infecting the stomachs of dogs and cats are potentially pathogenic and have been isolated from patients with gastric diseases. In the present study conducted in Japan, among the nine Helicobacter strains that we isolated from dogs and cats, NHP19-003T from a dog, and strains NHP19-012T and NHP21-005T from cats were identified to be the strains most closely related to Helicobacter heilmannii ASB1T based on a 16S rRNA comparison (98.7-99.2% similarity with H. heilmannii ASB1T). However, none of their whole genomes showed more than average nucleotide identity (ANI) threshold value (95-96%) to any Helicobacter species (85.1, 86.7, and 86.6% ANI, respectively, with H. heilmannii ASB1T), including when compared to each other. Furthermore, NHP19-003T, NHP19-012T, and NHP21-005T exhibited protein profiles different from known gastric Helicobacter species, as revealed by MALDI-TOF MS, indicating that they are novel Helicobacter species. We, thus, propose these novel Helicobacter species as follows: Helicobacter gastrocanis sp. nov. (type strain NHP19-003T [=JCM 39159T = DSM 111619T]), Helicobacter gastrofelis sp. nov. (type strain NHP19-012T [=JCM 39160T]) and Helicobacter felistomachi sp. nov. (type strain NHP21-005T [=JCM 39513T]). These novel strains have respective GC content values of 48.3, 46.9, and 47.1%. Phylogenetic analysis based on ureAB gene sequences obtained from gastric specimens from 47 dogs and 24 cats in Japan revealed that 29.8% of dogs were infected with H. gastrocanis, while H. gastrofelis infected 44.7% of dogs and 12.5% of cats. Additionally, 10.6% of dogs and 20.8% of cats were infected with H. felistomachi. Animal experiments have confirmed that these three novel species elicit gastric inflammatory responses. This study findings reveal the prevalence of novel gastric Helicobacter species in dogs and cats in Japan and their pathogenicity.
Keywords: cats; dogs; gastric Helicobacter species; gastric disease; novel species.
Copyright © 2025 Rimbara, Aoki, Suzuki, Kobayashi, Nakagawa, Goto-Koshino, Nomura, Du, Matsui, Mori, Shibayama, Kenri and Ohno.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Dewhirst F. E., Shen Z., Scimeca M. S., Stokes L. N., Boumenna T., Chen T., et al. (2005). Discordant 16S and 23S rRNA gene phylogenies for the genus Helicobacter: implications for phylogenetic inference and systematics. J. Bacteriol. 187, 6106–6118. doi: 10.1128/jb.187.17.6106-6118.2005, PMID: - DOI - PMC - PubMed
-
- Goodwin C. S., Armstrong J. A., Chilvers T., Peters M., Collins M. D., Sly L., et al. (1989). Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. Nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb, nov., respectively. Int. J. Syst. Evol. Microbiol. 39, 397–405. doi: 10.1099/00207713-39-4-397, PMID: - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

