This is a preprint.
Allosteric Modulation of Pathological Ataxin-3 Aggregation: A Path to Spinocerebellar Ataxia Type-3 Therapies
- PMID: 39896516
- PMCID: PMC11785186
- DOI: 10.1101/2025.01.22.633970
Allosteric Modulation of Pathological Ataxin-3 Aggregation: A Path to Spinocerebellar Ataxia Type-3 Therapies
Update in
-
Allosteric Modulation of Pathological Ataxin-3 Aggregation: A Path to Spinocerebellar Ataxia Type-3 Therapies.Adv Sci (Weinh). 2025 Nov 26:e02216. doi: 10.1002/advs.202502216. Online ahead of print. Adv Sci (Weinh). 2025. PMID: 41306023
Abstract
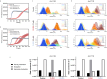
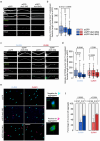
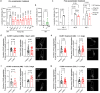
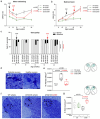
Spinocerebellar ataxia type 3 (SCA3) is a rare inherited neurodegenerative disease caused by the expansion of a polyglutamine repeat in the protease ataxin-3 (Atx3). Despite extensive knowledge of the downstream pathophysiology, no disease-modifying therapies are currently available to halt disease progression. The accumulation of protein inclusions enriched in the polyQ-expanded Atx3 in neurons suggests that inhibiting its self-assembly may yield targeted therapeutic approaches. Here it is shown that a supramolecular tweezer, CLR01, binds to a lysine residue on a positively charged surface patch of the Atx3 catalytic Josephin domain. At this site, the binding of CLR01 decreases the conformational fluctuations of the distal flexible hairpin. This results in reduced exposure of the nearby aggregation-prone region, which overlaps with the substrate ubiquitin binding site and primes Atx3 self-assembly, ultimately delaying Atx3 amyloid fibril formation and reducing the secondary nucleation rate, a process linked to fibril proliferation and toxicity. These effects translate into the reversal of synapse loss in a SCA3 cultured cortical neuron model, an improved locomotor function in a C. elegans SCA3 model, and a delay in disease onset, accompanied by reduced severity of motor symptoms in a SCA3 mouse model. This study provides critical insights into Atx3 self-assembly, revealing a novel allosteric site for designing CLR01-inspired therapies targeting pathological aggregation pathways while sparing essential functional sites. These findings emphasize that targeting allosteric sites in amyloid-forming proteins may offer unique opportunities to develop safe therapeutic strategies for various protein misfolding disorders.
Keywords: Amyloid; Molecular therapies; Polyglutamine; Pre-clinical models; Protein dynamics.
Figures


References
-
- Sicorello A., et al. , Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3. Structure, 2021. 29(1): p. 70–81 e5. - PubMed
-
- Seidel K., et al. , Brain pathology of spinocerebellar ataxias. Acta Neuropathol, 2012. 124(1): p. 1–21. - PubMed
-
- Schmidt J., et al. , Vulnerability of frontal brain neurons for the toxicity of expanded ataxin-3. Hum Mol Genet, 2019. 28(9): p. 1463–1473. - PubMed
