This is a preprint.
Construction of Multi-Modal Transcriptome-Small Molecule Interaction Networks from High-Throughput Measurements to Study Human Complex Traits
- PMID: 39896668
- PMCID: PMC11785221
- DOI: 10.1101/2025.01.22.634403
Construction of Multi-Modal Transcriptome-Small Molecule Interaction Networks from High-Throughput Measurements to Study Human Complex Traits
Abstract
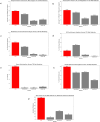
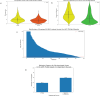
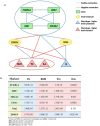
Small molecules (SMs) are integral to biological processes, influencing metabolism, homeostasis, and regulatory networks. Despite their importance, a significant knowledge gap exists regarding their downstream effects on biological pathways and gene expression, largely due to differences in scale, variability, and noise between untargeted metabolomics and sequencing-based technologies. To address these challenges, we developed a multi-omics framework comprising a machine learning-based protocol for data processing, a semi-supervised network inference approach, and network-guided analysis of complex traits. The ML protocol harmonized metabolomic, lipidomic, and transcriptomic data through batch correction, principal component analysis, and regression-based adjustments, enabling unbiased and effective integration. Building on this, we proposed a semi-supervised method to construct transcriptome-SM interaction networks (TSI-Nets) by selectively integrating SM profiles into gene-level networks using a meta-analytic approach that accounts for scale differences and missing data across omics layers. Benchmarking against three conventional unsupervised methods demonstrated the superiority of our approach in generating diverse, biologically relevant, and robust networks. While single-omics analyses identified 18 significant genes and 3 significant SMs associated with insulin sensitivity (IS), network-guided analysis revealed novel connections between these markers. The top-ranked module highlighted a cross-talk between fiber-degrading gut microbiota and immune regulatory pathways, inferred by the interaction of the protective SM, N-acetylglycine (NAG), with immune genes (FCER1A, HDC, MS4A2, and CPA3), linked to improved IS and reduced obesity and inflammation. Together, this framework offers a robust and scalable solution for multi-modal network inference and analysis, advancing SM pathway discovery and their implications for human health. Leveraging data from a population of thousands of individuals with extended longevity, the inferred TSI-Nets demonstrate generalizability across diverse conditions and complex traits. These networks are publicly available as a resource for the research community.
Conflict of interest statement
Declaration of Competing interest G.J.P. is a scientific advisory board member for Cambridge Isotope Laboratories and has a collaborative research agreement with Agilent Technologies. G.J.P. is the Chief Scientific Officer of Panome Bio.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
