This is a preprint.
Extensive genome evolution distinguishes maize within a stable tribe of grasses
- PMID: 39896679
- PMCID: PMC11785232
- DOI: 10.1101/2025.01.22.633974
Extensive genome evolution distinguishes maize within a stable tribe of grasses
Abstract
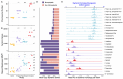
Over the last 20 million years, the Andropogoneae tribe of grasses has evolved to dominate 17% of global land area. Domestication of these grasses in the last 10,000 years has yielded our most productive crops, including maize, sugarcane, and sorghum. The majority of Andropogoneae species, including maize, show a history of polyploidy - a condition that, while offering the evolutionary advantage of multiple gene copies, poses challenges to basic cellular processes, gene expression, and epigenetic regulation. Genomic studies of polyploidy have been limited by sparse sampling of taxa in groups with multiple polyploidy events. Here, we present 33 genome assemblies from 27 species, including chromosome-scale assemblies of maize relatives Zea and Tripsacum. In maize, the after-effects of polyploidy have been widely studied, showing reduced chromosome number, biased fractionation of duplicate genes, and transposable element (TE) expansions. While we observe these patterns within the genus Zea, 12 other polyploidy events deviate significantly. Those tetraploids and hexaploids retain elevated chromosome number, maintain nearly complete complements of duplicate genes, and have only stochastic TE amplifications. These genomes reveal variable outcomes of polyploidy, challenging simple predictions and providing a foundation for understanding its evolutionary implications in an ecologically and economically important clade.
Figures




References
-
- Alexa A., & Rahnenführer J. (2009). Gene set enrichment analysis with topGO. Bioconductor Improv, 27, 1–26.
-
- Armstrong J., Hickey G., Diekhans M., Fiddes I. T., Novak A. M., Deran A., Fang Q., Xie D., Feng S., Stiller J., Genereux D., Johnson J., Marinescu V. D., Alföldi J., Harris R. S., Lindblad-Toh K., Haussler D., Karlsson E., Jarvis E. D., … Paten B. (2020). Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature, 587(7833), 246–251. 10.1038/s41586-020-2871-y - DOI - PMC - PubMed
-
- AuBuchon-Elder T., Minx P., Bookout B., & Kellogg E. A. (2023). Plant conservation assessment at scale: Rapid triage of extinction risks. PLANTS, PEOPLE, PLANET, 5(3), 386–397. 10.1002/ppp3.10355 - DOI
-
- Baduel P., Bray S., Vallejo-Marin M., Kolář F., & Yant L. (2018). The “Polyploid Hop”: Shifting Challenges and Opportunities Over the Evolutionary Lifespan of Genome Duplications. Frontiers in Ecology and Evolution, 6. 10.3389/fevo.2018.00117 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
