Enhancement of host defense against Helicobacter pylori infection through modulation of the gastrointestinal microenvironment by Lactiplantibacillus plantarum Lp05
- PMID: 39896799
- PMCID: PMC11782045
- DOI: 10.3389/fimmu.2024.1469885
Enhancement of host defense against Helicobacter pylori infection through modulation of the gastrointestinal microenvironment by Lactiplantibacillus plantarum Lp05
Abstract
Objective: This study aimed to assess the impact of Lactiplantibacillus plantarum Lp05 (Lp05) on the gastrointestinal microbiome and pathophysiological status of mice infected with Helicobacter pylori (H. pylori), exploring its potential as a probiotic treatment for H. pylori infections.
Methods: In vitro, the interaction between Lp05 and H. pylori was analyzed using laser confocal and scanning electron microscopy. In vivo, C57BL/6 mice infected with H. pylori were treated with Lp05 and divided into six groups: control, model, quadruple therapy, and three dosage levels of Lp05 (2×107, 2×108, 2×109 CFU/mouse/day). Over six weeks, the impact of Lp05 on the gastrointestinal microbiome and physiological markers was assessed. Measurements included digestive enzymes (α-amylase, pepsin, cellulase), inflammatory markers (interleukin-17A, interleukin-23, interleukin-10, interferon-β, interferon-γ, FoxP3, endothelin, IP-10, TGF-β1), oxidative stress markers (catalase, malondialdehyde, superoxide dismutase, myeloperoxidase), and tissue pathology (via modified Warthin-Starry silver and H&E staining). Microbial community structure in the stomach and intestines was evaluated through 16S rRNA gene sequencing.
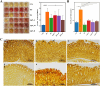
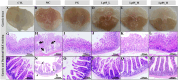
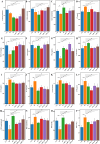
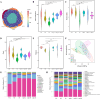
Results: In vitro studies showed Lp05 and H. pylori formed co-aggregates, with Lp05 potentially disrupting H. pylori cell structure, reducing its stomach colonization. In vivo, Lp05 significantly lowered gastric mucosal urease activity and serum H. pylori-IgG antibody levels in infected mice (p < 0.01). It also mitigated pathological changes in the stomach and duodenum, decreased inflammatory responses (ET, IL-17A, IL-23, TGF-beta1, and IP-10, p < 0.01 for all), and enhanced antioxidant enzyme activities (CAT and SOD, p < 0.01) while reducing MDA and MPO levels (p < 0.01), combating oxidative stress from H. pylori infection. Lp05 treatment significantly modified the intestinal and gastric microbiota, increasing beneficial bacteria like Lactobacillus and Ligilactobacillus, and decreasing harmful bacteria such as Olsenella, linked to pathological conditions.
Conclusion: Lp05 effectively modulates the gastrointestinal microbiome, reduces inflammation and oxidative stress, and suppresses H. pylori, promising for probiotic therapies with further research needed to refine its clinical use.
Keywords: Helicobacter pylori; Lactiplantibacillus plantarum; gastric microenvironment; gut microbiota; inflammatory markers.
Copyright © 2025 Dong, Han, Qi, Wu, Zhou, Jiang and Gai.
Conflict of interest statement
Authors YD, YQ and ZG were employed by Wecare Probiotics Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- FitzGerald R, Smith SM. An overview of helicobacter pylori infection. In: Smith SM, editor. Helicobacter pylori. Springer US, New York, NY: (2021). p. 1–14. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

