GEMCAT-a new algorithm for gene expression-based prediction of metabolic alterations
- PMID: 39897103
- PMCID: PMC11783570
- DOI: 10.1093/nargab/lqaf003
GEMCAT-a new algorithm for gene expression-based prediction of metabolic alterations
Abstract
The interpretation of multi-omics datasets obtained from high-throughput approaches is important to understand disease-related physiological changes and to predict biomarkers in body fluids. We present a new metabolite-centred genome-scale metabolic modelling algorithm, the Gene Expression-based Metabolite Centrality Analysis Tool (GEMCAT). GEMCAT enables integration of transcriptomics or proteomics data to predict changes in metabolite concentrations, which can be verified by targeted metabolomics. In addition, GEMCAT allows to trace measured and predicted metabolic changes back to the underlying alterations in gene expression or proteomics and thus enables functional interpretation and integration of multi-omics data. We demonstrate the predictive capacity of GEMCAT on three datasets and genome-scale metabolic networks from two different organisms: (i) we integrated transcriptomics and metabolomics data from an engineered human cell line with a functional deletion of the mitochondrial NAD transporter; (ii) we used a large multi-tissue multi-omics dataset from rats for transcriptome- and proteome-based prediction and verification of training-induced metabolic changes and achieved an average prediction accuracy of 70%; and (iii) we used proteomics measurements from patients with inflammatory bowel disease and verified the predicted changes using metabolomics data from the same patients. For this dataset, the prediction accuracy achieved by GEMCAT was 79%.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Conflict of interest statement
None declared.
Figures

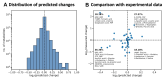

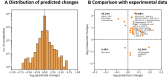

References
-
- Nielsen J. Systems biology of metabolism. Annu Rev Biochem. 2017; 86:245–75. - PubMed
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

