Innovative CDR grafting and computational methods for PD-1 specific nanobody design
- PMID: 39897125
- PMCID: PMC11782559
- DOI: 10.3389/fbinf.2024.1488331
Innovative CDR grafting and computational methods for PD-1 specific nanobody design
Abstract
Introduction: The development of nanobodies targeting Programmed Cell Death Protein-1 (PD-1) offers a promising approach in cancer immunotherapy. This study aims to design and characterize a PD-1-specific nanobody using an integrated computational and experimental approach.
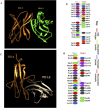
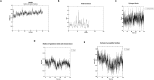
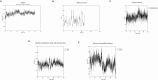
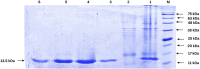
Methods: An in silico design strategy was employed, involving Complementarity-Determining Region (CDR) grafting to construct the nanobody sequence. The three-dimensional structure of the nanobody was predicted using AlphaFold2, and molecular docking simulations via ClusPro were conducted to evaluate binding interactions with PD-1. Physicochemical properties, including stability and solubility, were analyzed using web-based tools, while molecular dynamics (MD) simulations assessed stability under physiological conditions. The nanobody was produced and purified using Ni-NTA chromatography, and experimental validation was performed through Western blotting, ELISA, and dot blot analysis.
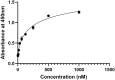
Results: Computational findings demonstrated favorable binding interactions, stability, and physicochemical properties of the nanobody. Experimental results confirmed the nanobody's specific binding affinity to PD-1, with ELISA and dot blot analyses providing evidence of robust interaction.
Discussion: This study highlights the potential of combining computational and experimental approaches for engineering nanobodies. The engineered PD-1 nanobody exhibits promising characteristics, making it a strong candidate for further testing in cancer immunotherapy applications.
Keywords: ELISA; Western blot; cancer immunotherapy; complementarity-determining region; dot blot; nanobody; programmed cell death protein-1.
Copyright © 2025 Devasani, Guntuku, Panatula, Muthyala, Palla and Siahaan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Design and Production of a Novel Anti-PD-1 Nanobody by CDR Grafting and Site-Directed Mutagenesis Approach.Mol Biotechnol. 2025 May;67(5):1843-1851. doi: 10.1007/s12033-024-01162-1. Epub 2024 May 12. Mol Biotechnol. 2025. PMID: 38736021
-
An integrated computational pipeline for designing high-affinity nanobodies with expanded genetic codes.Brief Bioinform. 2021 Nov 5;22(6):bbab338. doi: 10.1093/bib/bbab338. Brief Bioinform. 2021. PMID: 34415295
-
CDR grafting and site-directed mutagenesis approach for the generation and affinity maturation of Anti-CD20 nanobody.Mol Biol Rep. 2024 Jun 14;51(1):751. doi: 10.1007/s11033-024-09684-2. Mol Biol Rep. 2024. PMID: 38874667
-
Structural and Computational Studies of the SARS-CoV-2 Spike Protein Binding Mechanisms with Nanobodies: From Structure and Dynamics to Avidity-Driven Nanobody Engineering.Int J Mol Sci. 2022 Mar 8;23(6):2928. doi: 10.3390/ijms23062928. Int J Mol Sci. 2022. PMID: 35328351 Free PMC article. Review.
-
Nanobody engineering: computational modelling and design for biomedical and therapeutic applications.FEBS Open Bio. 2025 Feb;15(2):236-253. doi: 10.1002/2211-5463.13850. Epub 2024 Jun 19. FEBS Open Bio. 2025. PMID: 38898362 Free PMC article. Review.
References
-
- Bidkar A., Thakur N., Bolshette J. D., Gogoi R. (2014). In-silico structural and functional analysis of hypothetical proteins of leptospira interrogans. Biochem. Pharmacol. 3 (136), 2167–0501. 10.4172/2167-0501.1000136 - DOI
-
- Chang A. Y., Chau V., Landas J. A., Pang Y. (2017). Preparation of calcium competent Escherichia coli and heat-shock transformation. JEMI methods 1 (22-25).
LinkOut - more resources
Full Text Sources
Research Materials

