Polyploid QTL-seq identified QTLs controlling potato flesh color and tuber starch phosphorus content in a plexity-dependent manner
- PMID: 39897666
- PMCID: PMC11780331
- DOI: 10.1270/jsbbs.24028
Polyploid QTL-seq identified QTLs controlling potato flesh color and tuber starch phosphorus content in a plexity-dependent manner
Abstract
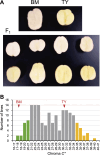
The progenies of polyploid crops inherit multiple sets of homoeologous chromosomes through various combinations, which impedes the identification of the quantitative trait loci (QTL) governing agronomic traits and the implementation of DNA marker-assisted breeding. Previously, we developed a whole-genome sequencing-based polyploid QTL-seq method that utilizes comprehensively extracted simplex polymorphisms for QTL mapping. Here, we verified the detection of duplex QTLs by modifying the analytical settings to explore the QTLs governing tuber flesh color and starch phosphorus content using tetraploid potato (Solanum tuberosum L.). The F1 progenies were obtained from a cross between 'Touya' (TY) and 'Benimaru' (BM). A single TY-derived QTL responsible for yellow flesh color was identified around a β-carotene hydroxylase gene on chromosome 3 using simplex polymorphisms, and a BM-derived QTL associated with decreased starch phosphorus content near a starch synthase II gene on chromosome 2 was detected using duplex polymorphisms. Furthermore, linked DNA markers were developed at the QTL sites. For the latter QTL, plexity-distinguishable markers were developed using quantitative PCR, fragment analysis, and amplicon sequencing. These revealed the allele dosage-dependent effect of the reduced starch phosphorus content. Thus, the polyploid QTL-seq pipeline can explore versatile QTLs beyond simplex, facilitating DNA marker-assisted breeding in various polyploid crops.
Keywords: DNA marker; QTL-seq; flesh color; plexity; polyploid; potato; starch phosphorus content.
Copyright © 2024 by JAPANESE SOCIETY OF BREEDING.
Figures



References
-
- Asano, K. and Tamiya S. (2016) Breeding of pest and disease resistant potato cultivars in Japan by using classical and molecular approaches. Jpn Agric Res Q 50: 1–6.
-
- Asano, K. and Endelman J.B. (2024) Development of KASP markers for the potato virus Y resistance gene Rychc using whole-genome resequencing data. Am J Potato Res 101: 114–121.

