Identification and validation of depression-associated genetic variants in the UK Biobank cohort with transcriptome and DNA methylation analyses in independent cohorts
- PMID: 39897774
- PMCID: PMC11787470
- DOI: 10.1016/j.heliyon.2025.e41865
Identification and validation of depression-associated genetic variants in the UK Biobank cohort with transcriptome and DNA methylation analyses in independent cohorts
Abstract
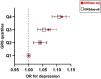
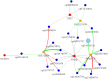
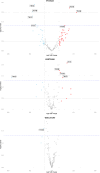
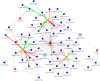
Depression is one of the most common psychiatric conditions resulting from a complex interaction of genetic, epigenetic and environmental factors. The present study aimed to identify independent genetic variants in the protein-coding genes that associate with depression and to analyze their transcriptomic and methylation profile. Data from the GWAS Catalogue was used to identify independent genetic variants for depression. The identified genetic variants were validated in the UK Biobank cohort and used to calculate a genetic risk score for depression. Data was also used from publicly available cohorts to conduct transcriptome and methylation analyses. Eight SNPs corresponding to six protein-coding genes (TNXB, NCAM1, LTBP3, BTN3A2, DAG1, FHIT) were identified that were highly associated with depression. These validated genetic variants for depression were used to calculate a genetic risk score that showed a significant association with depression (p < 0.05) but not with co-morbid traits. The transcriptome and methylation analyses suggested nominal significance for some gene probes (TNXB- and NCAM1) with depressed phenotype. The present study identified six protein-coding genes associated with depression and primarily involved in inflammation (TNXB), neuroplasticity (NCAM1 and LTBP3), immune response (BTN3A2), cell survival (DAG1) and circadian clock modification (FHIT). Our findings confirmed previous evidence for TNXB- and NCAM1 in the pathophysiology of depression and suggested new potential candidate genes (LTBP3, BTN3A2, DAG1 and FHIT) that warrant further investigation.
Keywords: Depression; Genetic risk score; Independent genetic variants; Methylation; Transcriptome; UK biobank.
© 2025 The Authors.
Conflict of interest statement
All authors declare no conflict of interest.
Figures

References
-
- McCarron R.M., Shapiro B., Rawles J., Luo J. Depression. Ann. Intern. Med. 2021;174:ITC65–ITC80. - PubMed
-
- Lang U.E., Borgwardt S. Molecular mechanisms of depression: perspectives on new treatment strategies. Cell. Physiol. Biochem. 2013;31:761–777. - PubMed
-
- Li Z., Page A., Martin G., Taylor R. Attributable risk of psychiatric and socio-economic factors for suicide from individual-level, population-based studies: a systematic review. Soc. Sci. Med. 2011;72:608–616. - PubMed
-
- Cavanagh J.T.O., Carson A.J., Sharpe M., Lawrie S.M. Psychological autopsy studies of suicide: a systematic review. Psychol. Med. 2003;33:395–405. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

