Transcriptome responses to Ralstonia solanacearum infection in tetraploid potato
- PMID: 39897796
- PMCID: PMC11786733
- DOI: 10.1016/j.heliyon.2025.e41903
Transcriptome responses to Ralstonia solanacearum infection in tetraploid potato
Abstract
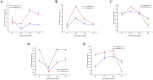
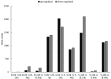
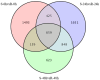
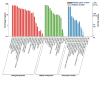
Potato (Solanum tuberosum) is an important global food source, the growth of which can be severely impacted by Ralstonia solanacearum bacterial infection. Despite extensive research, the molecular mechanisms of potato resistance to this pathogen are imperfectly known. Huashu No. 12, a tetraploid potato genotype, is highly resistant to R. solanacearum. We inoculate Huashu No. 12 and Longshu No. 7 (highly susceptible to R. solanacearum) with R. solanacearum to compare disease resistance in these two potato varieties. Huashu No. 12 has significantly higher resistance to R. solanacearum infection than Longshu No. 7, with increased lignin content, and an abundance of callose and strong autofluorescence in the phloem sieve tube. Enzymes (e.g., superoxide dismutase, catalase, peroxidase, phenylalanine ammonia-lyase, and polyphenol oxidase) contribute to R. solanacearum resistance in Huashu No. 12. Transcriptome sequencing reveals 659 differentially expressed genes between the two varieties, with the ethylene responsive factor family containing the most differentially expressed genes. Gene ontology and KEGG analyses provided further insights into the genetic basis and molecular mechanisms underlying plant defense against R. solanacearum disease. By demonstrating the importance of enzymes and differential gene expression in Huashu No. 12 resistance to R. solanacearum infection, the breeding of disease-resistant potato becomes increasingly feasible.
Keywords: Differentially expressed genes; Disease resistance; Potato; RNA-Seq; Ralstonia solanacearum.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Global transcriptome analyses reveal the molecular signatures in the early response of potato (Solanum tuberosum L.) to Phytophthora infestans, Ralstonia solanacearum, and Potato virus Y infection.Planta. 2020 Sep 21;252(4):57. doi: 10.1007/s00425-020-03471-6. Planta. 2020. PMID: 32955625
-
Transcriptome responses to Ralstonia solanacearum infection in the roots of the wild potato Solanum commersonii.BMC Genomics. 2015 Mar 26;16(1):246. doi: 10.1186/s12864-015-1460-1. BMC Genomics. 2015. PMID: 25880642 Free PMC article.
-
Interspecific Potato Breeding Lines Display Differential Colonization Patterns and Induced Defense Responses after Ralstonia solanacearum Infection.Front Plant Sci. 2017 Aug 28;8:1424. doi: 10.3389/fpls.2017.01424. eCollection 2017. Front Plant Sci. 2017. PMID: 28894453 Free PMC article.
-
Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era.Mol Plant Pathol. 2013 Sep;14(7):651-62. doi: 10.1111/mpp.12038. Epub 2013 May 30. Mol Plant Pathol. 2013. PMID: 23718203 Free PMC article. Review.
-
Strategies utilized by plants to defend against Ralstonia solanacearum.Front Plant Sci. 2025 May 26;16:1510177. doi: 10.3389/fpls.2025.1510177. eCollection 2025. Front Plant Sci. 2025. PMID: 40491817 Free PMC article. Review.
References
-
- Liu Y., et al. The sequevar distribution of Ralstonia solanacearum in tobacco-growing zones of China is structured by elevation. Eur. J. Plant Pathol. 2017;147(3):541–551.
-
- Deng R., et al. Progresses of main diseases resistance breeding in potato. Southwest China J. Agric. Sci. 2014;27(3):1337–1342.
LinkOut - more resources
Full Text Sources

