Low dimensionality of phenotypic space as an emergent property of coordinated teams in biological regulatory networks
- PMID: 39898023
- PMCID: PMC11787609
- DOI: 10.1016/j.isci.2024.111730
Low dimensionality of phenotypic space as an emergent property of coordinated teams in biological regulatory networks
Abstract
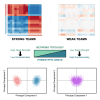
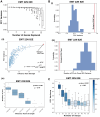
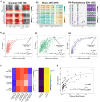
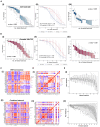
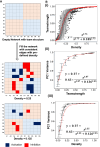
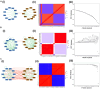
Cell-fate decisions involve coordinated genome-wide expression changes, typically leading to a limited number of phenotypes. Although often modeled as simple toggle switches, these rather simplistic representations often disregard the complexity of regulatory networks governing these changes. Here, we unravel design principles underlying complex cell decision-making networks in multiple contexts. We show that the emergent dynamics of these networks and corresponding transcriptomic data are consistently low-dimensional, as quantified by the variance explained by principal component 1 (PC1). This low dimensionality in phenotypic space arises from extensive feedback loops in these networks arranged to effectively enable the formation of two teams of mutually inhibiting nodes. We use team strength as a metric to quantify these feedback interactions and show its strong correlation with PC1 variance. Using artificial networks of varied topologies, we also establish the conditions for generating canalized cell-fate landscapes, offering insights into diverse binary cellular decision-making networks.
Keywords: Systems biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing financial or non-financial interests.
Figures


References
-
- Tabula Sapiens Consortium, Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P., et al. The Tabula Sapiens: A multiple- organ, single-cell transcriptomic atlas of humans. Science. 2022;376 doi: 10.1126/science.abl4896.eprint. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

